|
chr2_+_10123171
Show fit
|
2.39 |
ENST00000615152.5
|
RRM2
|
ribonucleotide reductase regulatory subunit M2
|
|
chr2_+_10122730
Show fit
|
2.15 |
ENST00000304567.10
|
RRM2
|
ribonucleotide reductase regulatory subunit M2
|
|
chr14_+_54397021
Show fit
|
1.91 |
ENST00000541304.5
|
CDKN3
|
cyclin dependent kinase inhibitor 3
|
|
chr14_+_54396949
Show fit
|
1.74 |
ENST00000611205.4
|
CDKN3
|
cyclin dependent kinase inhibitor 3
|
|
chr14_+_54396964
Show fit
|
1.72 |
ENST00000543789.6
ENST00000442975.6
ENST00000458126.6
ENST00000556102.6
ENST00000335183.11
|
CDKN3
|
cyclin dependent kinase inhibitor 3
|
|
chr4_+_17810945
Show fit
|
1.53 |
ENST00000251496.7
|
NCAPG
|
non-SMC condensin I complex subunit G
|
|
chr22_+_31082860
Show fit
|
1.49 |
ENST00000619644.4
|
SMTN
|
smoothelin
|
|
chr2_+_102311502
Show fit
|
1.43 |
ENST00000404917.6
ENST00000410040.5
|
IL1RL1
IL18R1
|
interleukin 1 receptor like 1
interleukin 18 receptor 1
|
|
chr16_+_56336727
Show fit
|
1.20 |
ENST00000568375.2
|
GNAO1
|
G protein subunit alpha o1
|
|
chr7_+_155298561
Show fit
|
1.08 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1
|
|
chr3_-_42702778
Show fit
|
1.04 |
ENST00000457462.5
ENST00000441594.6
|
HHATL
|
hedgehog acyltransferase like
|
|
chr1_-_24143112
Show fit
|
0.96 |
ENST00000270800.2
|
IL22RA1
|
interleukin 22 receptor subunit alpha 1
|
|
chr10_-_88952763
Show fit
|
0.95 |
ENST00000224784.10
|
ACTA2
|
actin alpha 2, smooth muscle
|
|
chr8_+_26577843
Show fit
|
0.92 |
ENST00000311151.9
|
DPYSL2
|
dihydropyrimidinase like 2
|
|
chr1_+_152908538
Show fit
|
0.90 |
ENST00000368764.4
|
IVL
|
involucrin
|
|
chr1_+_152514474
Show fit
|
0.90 |
ENST00000368790.4
|
CRCT1
|
cysteine rich C-terminal 1
|
|
chr10_-_73641450
Show fit
|
0.90 |
ENST00000359322.5
|
MYOZ1
|
myozenin 1
|
|
chr12_-_95548213
Show fit
|
0.86 |
ENST00000537435.2
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr19_-_11155798
Show fit
|
0.86 |
ENST00000592540.5
|
SPC24
|
SPC24 component of NDC80 kinetochore complex
|
|
chr1_-_153057504
Show fit
|
0.83 |
ENST00000392653.3
|
SPRR2A
|
small proline rich protein 2A
|
|
chr22_+_31081310
Show fit
|
0.82 |
ENST00000426927.5
ENST00000482444.5
ENST00000440425.5
ENST00000333137.12
ENST00000358743.5
ENST00000347557.6
|
SMTN
|
smoothelin
|
|
chr13_-_60013178
Show fit
|
0.81 |
ENST00000498416.2
ENST00000465066.5
|
DIAPH3
|
diaphanous related formin 3
|
|
chr3_+_122077850
Show fit
|
0.76 |
ENST00000482356.5
ENST00000393627.6
|
CD86
|
CD86 molecule
|
|
chr10_+_86958557
Show fit
|
0.73 |
ENST00000372017.4
ENST00000348795.8
|
SNCG
|
synuclein gamma
|
|
chr6_+_24774925
Show fit
|
0.71 |
ENST00000356509.7
ENST00000230056.8
|
GMNN
|
geminin DNA replication inhibitor
|
|
chr3_+_122077776
Show fit
|
0.70 |
ENST00000264468.9
|
CD86
|
CD86 molecule
|
|
chr12_-_95548837
Show fit
|
0.67 |
ENST00000393091.6
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr3_+_122055355
Show fit
|
0.67 |
ENST00000330540.7
ENST00000469710.5
ENST00000493101.5
|
CD86
|
CD86 molecule
|
|
chr19_+_676385
Show fit
|
0.67 |
ENST00000166139.9
|
FSTL3
|
follistatin like 3
|
|
chr2_-_55296361
Show fit
|
0.66 |
ENST00000647547.1
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr6_+_24775413
Show fit
|
0.66 |
ENST00000378054.6
ENST00000620958.4
ENST00000476555.5
|
GMNN
|
geminin DNA replication inhibitor
|
|
chr14_+_64704380
Show fit
|
0.64 |
ENST00000247226.13
ENST00000394691.7
|
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3
|
|
chr19_+_46601237
Show fit
|
0.64 |
ENST00000597743.5
|
CALM3
|
calmodulin 3
|
|
chr1_-_205449924
Show fit
|
0.63 |
ENST00000367154.5
|
LEMD1
|
LEM domain containing 1
|
|
chr9_+_35673917
Show fit
|
0.63 |
ENST00000617161.1
ENST00000378357.9
|
CA9
|
carbonic anhydrase 9
|
|
chr13_+_108596152
Show fit
|
0.63 |
ENST00000356711.7
ENST00000251041.10
|
MYO16
|
myosin XVI
|
|
chr12_-_98644733
Show fit
|
0.62 |
ENST00000299157.5
ENST00000393042.3
|
IKBIP
|
IKBKB interacting protein
|
|
chr12_+_48978313
Show fit
|
0.59 |
ENST00000293549.4
|
WNT1
|
Wnt family member 1
|
|
chr8_+_31640358
Show fit
|
0.59 |
ENST00000523534.5
|
NRG1
|
neuregulin 1
|
|
chr2_-_79087986
Show fit
|
0.56 |
ENST00000305089.8
|
REG1B
|
regenerating family member 1 beta
|
|
chr1_-_204166334
Show fit
|
0.55 |
ENST00000272190.9
|
REN
|
renin
|
|
chr15_-_82262660
Show fit
|
0.55 |
ENST00000557844.1
ENST00000359445.7
ENST00000268206.12
|
EFL1
|
elongation factor like GTPase 1
|
|
chr8_+_27325516
Show fit
|
0.55 |
ENST00000346049.10
ENST00000420218.3
|
PTK2B
|
protein tyrosine kinase 2 beta
|
|
chr17_-_78903193
Show fit
|
0.54 |
ENST00000322630.3
ENST00000586713.5
|
CEP295NL
|
CEP295 N-terminal like
|
|
chr4_-_164383986
Show fit
|
0.53 |
ENST00000507270.5
ENST00000514618.5
ENST00000503008.5
|
MARCHF1
|
membrane associated ring-CH-type finger 1
|
|
chr1_-_16978276
Show fit
|
0.51 |
ENST00000375534.7
|
MFAP2
|
microfibril associated protein 2
|
|
chr17_+_64449037
Show fit
|
0.51 |
ENST00000615220.4
ENST00000619286.5
ENST00000612535.4
ENST00000616498.4
|
MILR1
|
mast cell immunoglobulin like receptor 1
|
|
chr2_-_215138603
Show fit
|
0.50 |
ENST00000272895.12
|
ABCA12
|
ATP binding cassette subfamily A member 12
|
|
chr17_-_41055211
Show fit
|
0.49 |
ENST00000542910.1
ENST00000398477.1
|
KRTAP2-2
|
keratin associated protein 2-2
|
|
chr8_-_27258414
Show fit
|
0.48 |
ENST00000523048.5
|
STMN4
|
stathmin 4
|
|
chr11_-_107858777
Show fit
|
0.48 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2
|
|
chr15_-_70763430
Show fit
|
0.47 |
ENST00000539319.5
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr3_-_58587033
Show fit
|
0.47 |
ENST00000447756.2
|
FAM107A
|
family with sequence similarity 107 member A
|
|
chr1_+_119414931
Show fit
|
0.46 |
ENST00000543831.5
ENST00000433745.5
ENST00000369416.4
|
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2
|
|
chr1_-_151790475
Show fit
|
0.46 |
ENST00000368825.7
ENST00000368823.5
ENST00000368824.8
ENST00000458431.6
ENST00000368827.10
ENST00000440583.6
|
TDRKH
|
tudor and KH domain containing
|
|
chr2_+_218568865
Show fit
|
0.45 |
ENST00000295701.9
|
CNOT9
|
CCR4-NOT transcription complex subunit 9
|
|
chr14_-_54489003
Show fit
|
0.45 |
ENST00000554908.5
ENST00000616146.4
|
GMFB
|
glia maturation factor beta
|
|
chr14_-_54488940
Show fit
|
0.45 |
ENST00000628554.2
ENST00000358056.8
|
GMFB
|
glia maturation factor beta
|
|
chr10_-_44385043
Show fit
|
0.44 |
ENST00000374426.6
ENST00000395794.2
ENST00000374429.6
ENST00000395793.7
ENST00000343575.10
ENST00000395795.5
|
CXCL12
|
C-X-C motif chemokine ligand 12
|
|
chrX_+_47080855
Show fit
|
0.44 |
ENST00000336169.3
|
RGN
|
regucalcin
|
|
chrX_+_152830991
Show fit
|
0.44 |
ENST00000432467.1
|
NSDHL
|
NAD(P) dependent steroid dehydrogenase-like
|
|
chr17_-_19386785
Show fit
|
0.43 |
ENST00000497081.6
|
MFAP4
|
microfibril associated protein 4
|
|
chr19_-_14090963
Show fit
|
0.43 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chr4_+_168631597
Show fit
|
0.43 |
ENST00000504519.5
ENST00000512127.5
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr19_-_19262792
Show fit
|
0.42 |
ENST00000291481.8
|
HAPLN4
|
hyaluronan and proteoglycan link protein 4
|
|
chr2_+_112645930
Show fit
|
0.42 |
ENST00000272542.8
|
SLC20A1
|
solute carrier family 20 member 1
|
|
chr3_+_63443076
Show fit
|
0.42 |
ENST00000295894.9
|
SYNPR
|
synaptoporin
|
|
chr3_+_63443306
Show fit
|
0.42 |
ENST00000472899.5
ENST00000479198.5
ENST00000460711.5
ENST00000465156.1
|
SYNPR
|
synaptoporin
|
|
chr10_-_101588126
Show fit
|
0.42 |
ENST00000339310.7
ENST00000299206.8
ENST00000413344.5
ENST00000429502.1
ENST00000430045.1
ENST00000370172.5
ENST00000370162.8
ENST00000628479.2
|
POLL
|
DNA polymerase lambda
|
|
chr1_+_171257930
Show fit
|
0.41 |
ENST00000354841.4
|
FMO1
|
flavin containing dimethylaniline monoxygenase 1
|
|
chr16_+_56336767
Show fit
|
0.40 |
ENST00000640469.1
|
GNAO1
|
G protein subunit alpha o1
|
|
chr1_+_18630839
Show fit
|
0.40 |
ENST00000420770.7
|
PAX7
|
paired box 7
|
|
chr10_+_75111476
Show fit
|
0.39 |
ENST00000671730.1
|
SAMD8
|
sterile alpha motif domain containing 8
|
|
chr17_-_7393404
Show fit
|
0.39 |
ENST00000575434.4
|
PLSCR3
|
phospholipid scramblase 3
|
|
chr19_+_46601296
Show fit
|
0.39 |
ENST00000598871.5
ENST00000291295.14
ENST00000594523.5
|
CALM3
|
calmodulin 3
|
|
chr21_-_15064934
Show fit
|
0.39 |
ENST00000400199.5
ENST00000400202.5
ENST00000318948.7
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr6_+_106098933
Show fit
|
0.38 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1
|
|
chr11_+_61248122
Show fit
|
0.38 |
ENST00000451616.6
|
PGA5
|
pepsinogen A5
|
|
chr10_+_75111595
Show fit
|
0.38 |
ENST00000671800.1
ENST00000542569.6
ENST00000372687.4
|
SAMD8
|
sterile alpha motif domain containing 8
|
|
chr16_+_56336805
Show fit
|
0.38 |
ENST00000564727.2
|
GNAO1
|
G protein subunit alpha o1
|
|
chr11_+_35139162
Show fit
|
0.38 |
ENST00000415148.6
ENST00000263398.11
ENST00000428726.8
ENST00000526669.6
ENST00000433892.6
ENST00000525211.6
ENST00000278386.10
ENST00000434472.6
ENST00000352818.8
ENST00000442151.6
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr1_-_77979054
Show fit
|
0.38 |
ENST00000370768.7
ENST00000370767.5
ENST00000421641.1
|
FUBP1
|
far upstream element binding protein 1
|
|
chr5_+_96662046
Show fit
|
0.37 |
ENST00000338252.7
ENST00000508830.5
|
CAST
|
calpastatin
|
|
chr11_+_62419025
Show fit
|
0.37 |
ENST00000278282.3
|
SCGB1A1
|
secretoglobin family 1A member 1
|
|
chr7_+_31687208
Show fit
|
0.37 |
ENST00000409146.3
ENST00000342032.8
|
PPP1R17
|
protein phosphatase 1 regulatory subunit 17
|
|
chr1_+_119507203
Show fit
|
0.37 |
ENST00000369413.8
ENST00000528909.1
|
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1
|
|
chr14_-_24141570
Show fit
|
0.37 |
ENST00000560403.5
ENST00000419198.6
ENST00000216799.9
|
EMC9
|
ER membrane protein complex subunit 9
|
|
chr11_+_8682782
Show fit
|
0.37 |
ENST00000531978.5
ENST00000524496.5
ENST00000532359.5
ENST00000314138.11
ENST00000530022.5
|
RPL27A
|
ribosomal protein L27a
|
|
chr3_-_28348629
Show fit
|
0.37 |
ENST00000334100.10
|
AZI2
|
5-azacytidine induced 2
|
|
chr7_+_24573415
Show fit
|
0.37 |
ENST00000409761.5
ENST00000222644.10
ENST00000396475.6
|
MPP6
|
membrane palmitoylated protein 6
|
|
chr2_-_223837553
Show fit
|
0.36 |
ENST00000396654.7
ENST00000396653.2
ENST00000443700.5
|
AP1S3
|
adaptor related protein complex 1 subunit sigma 3
|
|
chr1_+_15756659
Show fit
|
0.36 |
ENST00000375771.5
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr3_+_14675128
Show fit
|
0.36 |
ENST00000435614.5
ENST00000253697.8
ENST00000412910.1
|
C3orf20
|
chromosome 3 open reading frame 20
|
|
chr16_+_30064142
Show fit
|
0.36 |
ENST00000562168.5
ENST00000569545.5
|
ALDOA
|
aldolase, fructose-bisphosphate A
|
|
chr1_+_156893678
Show fit
|
0.35 |
ENST00000292357.8
ENST00000338302.7
ENST00000455314.5
|
PEAR1
|
platelet endothelial aggregation receptor 1
|
|
chr21_-_37267511
Show fit
|
0.35 |
ENST00000398998.1
|
VPS26C
|
VPS26 endosomal protein sorting factor C
|
|
chr16_-_138512
Show fit
|
0.35 |
ENST00000399953.7
|
NPRL3
|
NPR3 like, GATOR1 complex subunit
|
|
chr5_+_132073782
Show fit
|
0.35 |
ENST00000296871.4
|
CSF2
|
colony stimulating factor 2
|
|
chr3_-_28348924
Show fit
|
0.34 |
ENST00000414162.5
ENST00000420543.6
|
AZI2
|
5-azacytidine induced 2
|
|
chr2_-_223837484
Show fit
|
0.34 |
ENST00000446015.6
ENST00000409375.1
|
AP1S3
|
adaptor related protein complex 1 subunit sigma 3
|
|
chr14_-_106374129
Show fit
|
0.34 |
ENST00000390616.2
|
IGHV4-34
|
immunoglobulin heavy variable 4-34
|
|
chrX_+_38352573
Show fit
|
0.34 |
ENST00000039007.5
|
OTC
|
ornithine transcarbamylase
|
|
chr16_-_3099228
Show fit
|
0.34 |
ENST00000572431.1
ENST00000572548.1
ENST00000575108.5
ENST00000576985.6
ENST00000576483.1
ENST00000538082.5
|
ZSCAN10
|
zinc finger and SCAN domain containing 10
|
|
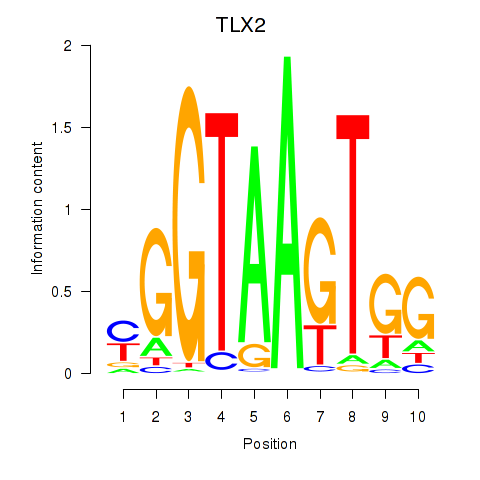
chr14_-_56805648
Show fit
|
0.34 |
ENST00000554788.5
ENST00000554845.1
ENST00000408990.8
|
OTX2
|
orthodenticle homeobox 2
|
|
chr19_-_45406327
Show fit
|
0.34 |
ENST00000593226.5
ENST00000418234.6
|
PPP1R13L
|
protein phosphatase 1 regulatory subunit 13 like
|
|
chr18_+_31318144
Show fit
|
0.34 |
ENST00000257192.5
|
DSG1
|
desmoglein 1
|
|
chr9_-_112175283
Show fit
|
0.33 |
ENST00000374270.8
ENST00000374263.7
|
SUSD1
|
sushi domain containing 1
|
|
chr8_+_38901327
Show fit
|
0.33 |
ENST00000519640.5
ENST00000617275.5
|
PLEKHA2
|
pleckstrin homology domain containing A2
|
|
chr9_+_123356189
Show fit
|
0.33 |
ENST00000373631.8
|
CRB2
|
crumbs cell polarity complex component 2
|
|
chr9_+_136952896
Show fit
|
0.33 |
ENST00000371632.7
|
LCN12
|
lipocalin 12
|
|
chr21_-_42350987
Show fit
|
0.32 |
ENST00000291526.5
|
TFF2
|
trefoil factor 2
|
|
chr19_+_2389761
Show fit
|
0.32 |
ENST00000648592.1
|
TMPRSS9
|
transmembrane serine protease 9
|
|
chr11_-_93741479
Show fit
|
0.32 |
ENST00000448108.7
ENST00000532455.1
|
TAF1D
|
TATA-box binding protein associated factor, RNA polymerase I subunit D
|
|
chr1_-_44843240
Show fit
|
0.32 |
ENST00000372192.4
|
PTCH2
|
patched 2
|
|
chr10_-_33334625
Show fit
|
0.32 |
ENST00000374875.5
ENST00000374822.8
ENST00000374867.7
|
NRP1
|
neuropilin 1
|
|
chr8_-_130443581
Show fit
|
0.32 |
ENST00000357668.2
ENST00000518721.6
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1
|
|
chr11_+_60455839
Show fit
|
0.32 |
ENST00000532491.5
ENST00000532073.5
ENST00000345732.9
ENST00000534668.6
ENST00000528313.1
ENST00000533306.6
ENST00000674194.1
|
MS4A1
|
membrane spanning 4-domains A1
|
|
chr1_+_86424154
Show fit
|
0.31 |
ENST00000370565.5
|
CLCA2
|
chloride channel accessory 2
|
|
chr11_+_18599782
Show fit
|
0.31 |
ENST00000634992.1
ENST00000635674.1
ENST00000511927.2
|
SPTY2D1OS
|
SPTY2D1 opposite strand
|
|
chr7_+_77840122
Show fit
|
0.31 |
ENST00000450574.5
ENST00000248550.7
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr6_-_43528867
Show fit
|
0.31 |
ENST00000455285.2
|
XPO5
|
exportin 5
|
|
chr8_+_79611727
Show fit
|
0.31 |
ENST00000518491.1
|
STMN2
|
stathmin 2
|
|
chr11_+_62856004
Show fit
|
0.31 |
ENST00000680729.1
|
SLC3A2
|
solute carrier family 3 member 2
|
|
chr5_+_96662314
Show fit
|
0.31 |
ENST00000674702.1
|
CAST
|
calpastatin
|
|
chr8_+_133191060
Show fit
|
0.31 |
ENST00000519433.1
ENST00000517423.5
ENST00000220856.6
|
CCN4
|
cellular communication network factor 4
|
|
chr7_-_100168602
Show fit
|
0.31 |
ENST00000423751.2
ENST00000360039.9
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr10_+_122271292
Show fit
|
0.31 |
ENST00000260723.6
|
BTBD16
|
BTB domain containing 16
|
|
chr1_+_52602347
Show fit
|
0.30 |
ENST00000361314.5
|
GPX7
|
glutathione peroxidase 7
|
|
chr18_+_57435366
Show fit
|
0.30 |
ENST00000491143.3
|
ONECUT2
|
one cut homeobox 2
|
|
chr11_-_83682414
Show fit
|
0.30 |
ENST00000404783.7
|
DLG2
|
discs large MAGUK scaffold protein 2
|
|
chr5_+_96662214
Show fit
|
0.30 |
ENST00000395812.6
|
CAST
|
calpastatin
|
|
chr8_-_104588998
Show fit
|
0.30 |
ENST00000424843.6
|
LRP12
|
LDL receptor related protein 12
|
|
chr10_+_62374361
Show fit
|
0.29 |
ENST00000395254.8
|
ZNF365
|
zinc finger protein 365
|
|
chr19_+_47713412
Show fit
|
0.29 |
ENST00000538399.1
ENST00000263277.8
|
EHD2
|
EH domain containing 2
|
|
chr9_+_136665745
Show fit
|
0.29 |
ENST00000371698.3
|
EGFL7
|
EGF like domain multiple 7
|
|
chr20_+_58309704
Show fit
|
0.29 |
ENST00000244040.4
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chrX_-_133218345
Show fit
|
0.29 |
ENST00000310125.5
|
TFDP3
|
transcription factor Dp family member 3
|
|
chr16_+_30064274
Show fit
|
0.29 |
ENST00000563060.6
|
ALDOA
|
aldolase, fructose-bisphosphate A
|
|
chrX_-_15664798
Show fit
|
0.29 |
ENST00000380342.4
|
CLTRN
|
collectrin, amino acid transport regulator
|
|
chr17_+_44847905
Show fit
|
0.29 |
ENST00000587021.1
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B
|
|
chr6_-_32666648
Show fit
|
0.29 |
ENST00000399082.7
ENST00000399079.7
ENST00000374943.8
ENST00000434651.6
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1
|
|
chr15_-_65106308
Show fit
|
0.29 |
ENST00000502113.2
|
UBAP1L
|
ubiquitin associated protein 1 like
|
|
chr12_+_56338873
Show fit
|
0.29 |
ENST00000228534.6
|
IL23A
|
interleukin 23 subunit alpha
|
|
chr5_+_96662366
Show fit
|
0.28 |
ENST00000675179.1
ENST00000421689.6
ENST00000674984.1
ENST00000512620.5
|
CAST
|
calpastatin
|
|
chr6_-_49744378
Show fit
|
0.28 |
ENST00000371159.8
ENST00000263045.9
|
CRISP3
|
cysteine rich secretory protein 3
|
|
chr2_-_287687
Show fit
|
0.28 |
ENST00000401489.6
ENST00000619265.4
|
ALKAL2
|
ALK and LTK ligand 2
|
|
chr6_-_49744434
Show fit
|
0.28 |
ENST00000433368.6
ENST00000354620.4
|
CRISP3
|
cysteine rich secretory protein 3
|
|
chr19_+_4402615
Show fit
|
0.28 |
ENST00000301280.10
|
CHAF1A
|
chromatin assembly factor 1 subunit A
|
|
chr16_+_30064462
Show fit
|
0.28 |
ENST00000412304.6
|
ALDOA
|
aldolase, fructose-bisphosphate A
|
|
chr9_-_71446835
Show fit
|
0.28 |
ENST00000357533.6
|
TRPM3
|
transient receptor potential cation channel subfamily M member 3
|
|
chr12_+_12891554
Show fit
|
0.28 |
ENST00000014914.6
|
GPRC5A
|
G protein-coupled receptor class C group 5 member A
|
|
chr5_+_149271848
Show fit
|
0.27 |
ENST00000296721.9
|
AFAP1L1
|
actin filament associated protein 1 like 1
|
|
chr1_+_149782671
Show fit
|
0.27 |
ENST00000444948.5
ENST00000369168.5
|
FCGR1A
|
Fc fragment of IgG receptor Ia
|
|
chr16_+_31108294
Show fit
|
0.27 |
ENST00000287507.7
ENST00000394950.7
ENST00000219794.11
ENST00000561755.1
|
BCKDK
|
branched chain keto acid dehydrogenase kinase
|
|
chr8_-_104589241
Show fit
|
0.27 |
ENST00000276654.10
|
LRP12
|
LDL receptor related protein 12
|
|
chr11_+_65576023
Show fit
|
0.27 |
ENST00000533237.5
ENST00000309295.9
ENST00000634639.1
|
EHBP1L1
|
EH domain binding protein 1 like 1
|
|
chr2_-_287299
Show fit
|
0.27 |
ENST00000405290.5
|
ALKAL2
|
ALK and LTK ligand 2
|
|
chr19_+_15641280
Show fit
|
0.27 |
ENST00000585846.1
|
CYP4F3
|
cytochrome P450 family 4 subfamily F member 3
|
|
chr2_-_287320
Show fit
|
0.26 |
ENST00000401503.5
|
ALKAL2
|
ALK and LTK ligand 2
|
|
chr12_+_71439976
Show fit
|
0.26 |
ENST00000536515.5
ENST00000540815.2
|
LGR5
|
leucine rich repeat containing G protein-coupled receptor 5
|
|
chr2_-_43838832
Show fit
|
0.26 |
ENST00000405322.8
|
ABCG5
|
ATP binding cassette subfamily G member 5
|
|
chrX_+_49053554
Show fit
|
0.26 |
ENST00000597275.5
|
CCDC120
|
coiled-coil domain containing 120
|
|
chr18_+_31376777
Show fit
|
0.26 |
ENST00000308128.9
ENST00000359747.4
|
DSG4
|
desmoglein 4
|
|
chr8_+_38901757
Show fit
|
0.26 |
ENST00000616834.1
|
PLEKHA2
|
pleckstrin homology domain containing A2
|
|
chr17_-_2266131
Show fit
|
0.26 |
ENST00000570606.5
ENST00000354901.8
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor
|
|
chr3_-_183162726
Show fit
|
0.26 |
ENST00000265598.8
|
LAMP3
|
lysosomal associated membrane protein 3
|
|
chr20_+_45207025
Show fit
|
0.26 |
ENST00000372781.4
|
SEMG1
|
semenogelin 1
|
|
chr1_+_31296989
Show fit
|
0.26 |
ENST00000373714.5
ENST00000344147.10
ENST00000627541.1
|
ZCCHC17
|
zinc finger CCHC-type containing 17
|
|
chr11_+_1839452
Show fit
|
0.25 |
ENST00000381906.5
|
TNNI2
|
troponin I2, fast skeletal type
|
|
chr4_+_119135825
Show fit
|
0.25 |
ENST00000307128.6
|
MYOZ2
|
myozenin 2
|
|
chr4_-_5019437
Show fit
|
0.25 |
ENST00000506508.1
ENST00000509419.1
ENST00000307746.9
|
CYTL1
|
cytokine like 1
|
|
chr5_+_95646854
Show fit
|
0.25 |
ENST00000311364.8
ENST00000458310.1
|
RFESD
|
Rieske Fe-S domain containing
|
|
chr18_+_31498168
Show fit
|
0.25 |
ENST00000261590.13
ENST00000585206.1
ENST00000683654.1
|
DSG2
|
desmoglein 2
|
|
chr2_-_203535253
Show fit
|
0.25 |
ENST00000457812.5
ENST00000319170.10
ENST00000630330.2
ENST00000308091.8
ENST00000453034.5
ENST00000420371.2
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr19_+_35292145
Show fit
|
0.25 |
ENST00000595791.5
ENST00000597035.5
ENST00000537831.2
ENST00000392213.8
|
MAG
|
myelin associated glycoprotein
|
|
chr12_-_16606795
Show fit
|
0.25 |
ENST00000447609.5
|
LMO3
|
LIM domain only 3
|
|
chr6_+_13272709
Show fit
|
0.24 |
ENST00000379335.8
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr3_+_46354072
Show fit
|
0.24 |
ENST00000445132.3
ENST00000421659.1
|
CCR2
|
C-C motif chemokine receptor 2
|
|
chr5_+_150497772
Show fit
|
0.24 |
ENST00000523767.5
|
NDST1
|
N-deacetylase and N-sulfotransferase 1
|
|
chr18_+_24460655
Show fit
|
0.24 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4
|
|
chr7_-_22500152
Show fit
|
0.24 |
ENST00000406890.6
ENST00000678116.1
ENST00000424363.5
|
STEAP1B
|
STEAP family member 1B
|
|
chr1_+_209938169
Show fit
|
0.24 |
ENST00000367019.5
ENST00000537238.5
ENST00000637265.1
|
SYT14
|
synaptotagmin 14
|
|
chr5_-_16916400
Show fit
|
0.24 |
ENST00000513882.5
|
MYO10
|
myosin X
|
|
chr17_-_7404039
Show fit
|
0.24 |
ENST00000576017.1
ENST00000302422.4
|
TMEM256
|
transmembrane protein 256
|
|
chr19_+_35292125
Show fit
|
0.24 |
ENST00000600291.5
ENST00000361922.8
|
MAG
|
myelin associated glycoprotein
|
|
chr18_+_24460630
Show fit
|
0.24 |
ENST00000256906.5
|
HRH4
|
histamine receptor H4
|
|
chr16_+_30000944
Show fit
|
0.24 |
ENST00000562291.2
|
INO80E
|
INO80 complex subunit E
|
|
chr7_-_11832190
Show fit
|
0.24 |
ENST00000423059.9
ENST00000617773.1
|
THSD7A
|
thrombospondin type 1 domain containing 7A
|
|
chr17_-_81166160
Show fit
|
0.23 |
ENST00000326724.9
|
AATK
|
apoptosis associated tyrosine kinase
|
|
chr5_+_96662969
Show fit
|
0.23 |
ENST00000514845.5
ENST00000675663.1
|
CAST
|
calpastatin
|
|
chr20_+_44355692
Show fit
|
0.23 |
ENST00000316673.8
ENST00000609795.5
ENST00000457232.5
ENST00000609262.5
|
HNF4A
|
hepatocyte nuclear factor 4 alpha
|
|
chr20_+_36214373
Show fit
|
0.23 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1
|
|
chr1_+_209938207
Show fit
|
0.23 |
ENST00000472886.5
|
SYT14
|
synaptotagmin 14
|
|
chr12_-_16606102
Show fit
|
0.23 |
ENST00000537304.6
|
LMO3
|
LIM domain only 3
|
|
chr7_+_139829242
Show fit
|
0.23 |
ENST00000455353.6
ENST00000458722.6
ENST00000448866.7
ENST00000411653.6
|
TBXAS1
|
thromboxane A synthase 1
|
|
chr5_-_160370619
Show fit
|
0.23 |
ENST00000652664.2
ENST00000393975.4
|
C1QTNF2
|
C1q and TNF related 2
|
|
chr1_-_21937300
Show fit
|
0.23 |
ENST00000374695.8
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr12_-_16605939
Show fit
|
0.23 |
ENST00000541295.5
ENST00000535535.5
|
LMO3
|
LIM domain only 3
|
|
chr1_+_155859550
Show fit
|
0.23 |
ENST00000368324.5
|
SYT11
|
synaptotagmin 11
|
|
chr21_-_37267300
Show fit
|
0.23 |
ENST00000309117.11
ENST00000476950.5
ENST00000399001.5
|
VPS26C
|
VPS26 endosomal protein sorting factor C
|