Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
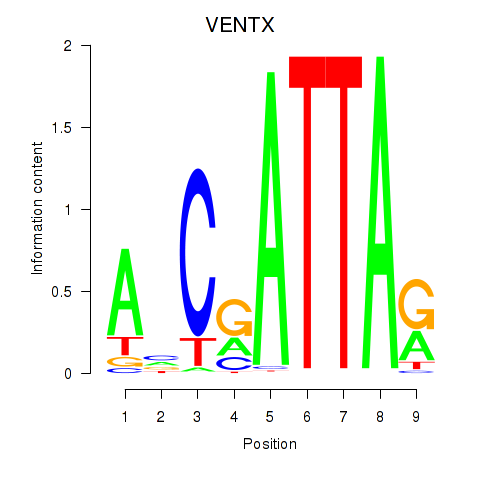
Results for VENTX
Z-value: 0.48
Transcription factors associated with VENTX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VENTX
|
ENSG00000151650.8 | VENTX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VENTX | hg38_v1_chr10_+_133237849_133237863 | -0.01 | 9.6e-01 | Click! |
Activity profile of VENTX motif
Sorted Z-values of VENTX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of VENTX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_32668368 | 1.07 |
ENST00000399084.5
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1 |
| chr13_+_108629605 | 0.93 |
ENST00000457511.7
|
MYO16
|
myosin XVI |
| chr1_+_244352627 | 0.79 |
ENST00000366537.5
ENST00000308105.5 |
C1orf100
|
chromosome 1 open reading frame 100 |
| chr6_-_39725335 | 0.70 |
ENST00000538893.5
|
KIF6
|
kinesin family member 6 |
| chr12_-_86256267 | 0.63 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr12_+_107318395 | 0.58 |
ENST00000420571.6
ENST00000280758.10 |
BTBD11
|
BTB domain containing 11 |
| chr1_+_174447944 | 0.51 |
ENST00000367685.5
|
GPR52
|
G protein-coupled receptor 52 |
| chr1_+_40247926 | 0.50 |
ENST00000372766.4
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr3_-_57226344 | 0.48 |
ENST00000495160.2
|
HESX1
|
HESX homeobox 1 |
| chr10_-_25062279 | 0.47 |
ENST00000615958.4
|
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr17_-_41047267 | 0.42 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr4_-_154590735 | 0.38 |
ENST00000403106.8
ENST00000622532.1 ENST00000651975.1 |
FGA
|
fibrinogen alpha chain |
| chr17_-_10838075 | 0.36 |
ENST00000580256.3
ENST00000643787.1 |
PIRT
ENSG00000284876.1
|
phosphoinositide interacting regulator of transient receptor potential channels novel transcript |
| chr6_-_49713564 | 0.35 |
ENST00000616725.4
ENST00000618917.4 |
CRISP2
|
cysteine rich secretory protein 2 |
| chr8_+_49911604 | 0.34 |
ENST00000642164.1
ENST00000644093.1 ENST00000643999.1 ENST00000647073.1 ENST00000646880.1 |
SNTG1
|
syntrophin gamma 1 |
| chr7_+_120988683 | 0.33 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr6_-_49713521 | 0.33 |
ENST00000339139.5
|
CRISP2
|
cysteine rich secretory protein 2 |
| chr4_-_26490453 | 0.33 |
ENST00000295589.4
|
CCKAR
|
cholecystokinin A receptor |
| chr8_+_49911396 | 0.30 |
ENST00000642720.2
|
SNTG1
|
syntrophin gamma 1 |
| chr14_+_64704380 | 0.28 |
ENST00000247226.13
ENST00000394691.7 |
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr14_+_32329341 | 0.28 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr2_-_162152404 | 0.27 |
ENST00000375497.3
|
GCG
|
glucagon |
| chr12_-_89352395 | 0.27 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr3_-_57199938 | 0.27 |
ENST00000473921.2
ENST00000295934.8 |
HESX1
|
HESX homeobox 1 |
| chr3_+_186666003 | 0.26 |
ENST00000232003.5
|
HRG
|
histidine rich glycoprotein |
| chr12_-_89352487 | 0.26 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr8_+_31639222 | 0.26 |
ENST00000519301.6
ENST00000652698.1 |
NRG1
|
neuregulin 1 |
| chr18_+_44700796 | 0.26 |
ENST00000677130.1
|
SETBP1
|
SET binding protein 1 |
| chr8_+_31639291 | 0.25 |
ENST00000651149.1
ENST00000650866.1 |
NRG1
|
neuregulin 1 |
| chr12_-_52434363 | 0.25 |
ENST00000252245.6
|
KRT75
|
keratin 75 |
| chr4_+_168497066 | 0.25 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_+_202073282 | 0.24 |
ENST00000459709.5
|
KIAA2012
|
KIAA2012 |
| chr12_-_119803383 | 0.24 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr7_-_28180735 | 0.24 |
ENST00000283928.10
|
JAZF1
|
JAZF zinc finger 1 |
| chr7_+_18495723 | 0.23 |
ENST00000681950.1
ENST00000622668.4 ENST00000405010.7 ENST00000406451.8 ENST00000441542.7 ENST00000428307.6 ENST00000681273.1 |
HDAC9
|
histone deacetylase 9 |
| chr1_+_153774210 | 0.23 |
ENST00000271857.6
|
SLC27A3
|
solute carrier family 27 member 3 |
| chr14_+_92323154 | 0.23 |
ENST00000532405.6
ENST00000676001.1 ENST00000531433.5 |
SLC24A4
|
solute carrier family 24 member 4 |
| chr4_+_128811311 | 0.23 |
ENST00000413543.6
|
JADE1
|
jade family PHD finger 1 |
| chr4_+_168497044 | 0.23 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_+_202073249 | 0.23 |
ENST00000498697.3
|
KIAA2012
|
KIAA2012 |
| chr6_-_75493773 | 0.22 |
ENST00000237172.12
|
FILIP1
|
filamin A interacting protein 1 |
| chr4_+_128811264 | 0.22 |
ENST00000610919.4
ENST00000510308.5 |
JADE1
|
jade family PHD finger 1 |
| chr2_-_207167220 | 0.22 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chrX_+_136648138 | 0.22 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr12_-_101210232 | 0.21 |
ENST00000536262.3
|
SLC5A8
|
solute carrier family 5 member 8 |
| chr12_-_119804298 | 0.21 |
ENST00000678652.1
ENST00000678494.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr6_-_75493629 | 0.21 |
ENST00000393004.6
|
FILIP1
|
filamin A interacting protein 1 |
| chr14_-_26597430 | 0.21 |
ENST00000344429.9
ENST00000574031.1 ENST00000465357.6 ENST00000547619.5 |
NOVA1
|
NOVA alternative splicing regulator 1 |
| chr8_+_49911801 | 0.20 |
ENST00000643809.1
|
SNTG1
|
syntrophin gamma 1 |
| chr1_+_240123121 | 0.20 |
ENST00000681210.1
|
FMN2
|
formin 2 |
| chr1_+_240123148 | 0.20 |
ENST00000681824.1
|
FMN2
|
formin 2 |
| chr17_-_7404039 | 0.20 |
ENST00000576017.1
ENST00000302422.4 |
TMEM256
|
transmembrane protein 256 |
| chr12_-_52618559 | 0.20 |
ENST00000305748.7
|
KRT73
|
keratin 73 |
| chr4_+_168092530 | 0.19 |
ENST00000359299.8
|
ANXA10
|
annexin A10 |
| chr8_-_86743626 | 0.19 |
ENST00000320005.6
|
CNGB3
|
cyclic nucleotide gated channel subunit beta 3 |
| chr1_-_101846957 | 0.18 |
ENST00000338858.9
|
OLFM3
|
olfactomedin 3 |
| chr16_-_49856105 | 0.18 |
ENST00000563137.7
|
ZNF423
|
zinc finger protein 423 |
| chr12_+_69825221 | 0.18 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like |
| chr5_+_62578810 | 0.18 |
ENST00000334994.6
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr14_+_32329256 | 0.18 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr12_+_69825273 | 0.17 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr2_+_176157293 | 0.17 |
ENST00000683222.1
|
HOXD3
|
homeobox D3 |
| chr4_-_137532452 | 0.17 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr6_-_132659178 | 0.17 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr11_-_107858777 | 0.17 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr7_-_83162899 | 0.16 |
ENST00000423517.6
|
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr6_-_161274042 | 0.16 |
ENST00000320285.9
|
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr14_+_64715677 | 0.16 |
ENST00000634379.2
|
PLEKHG3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr6_-_161274010 | 0.16 |
ENST00000366911.9
ENST00000366905.3 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr7_-_151210488 | 0.15 |
ENST00000644661.2
|
H2BE1
|
H2B.E variant histone 1 |
| chr11_-_27722021 | 0.14 |
ENST00000314915.6
|
BDNF
|
brain derived neurotrophic factor |
| chr7_-_78771265 | 0.14 |
ENST00000630991.2
ENST00000629359.2 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr20_-_52105644 | 0.14 |
ENST00000371523.8
|
ZFP64
|
ZFP64 zinc finger protein |
| chrX_+_136648214 | 0.14 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr5_-_80654956 | 0.14 |
ENST00000439211.7
|
DHFR
|
dihydrofolate reductase |
| chr10_+_112283399 | 0.14 |
ENST00000643850.1
ENST00000646139.2 ENST00000645243.1 |
TECTB
|
tectorin beta |
| chr12_-_56934403 | 0.13 |
ENST00000293502.2
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C member 7 |
| chr12_+_111034136 | 0.12 |
ENST00000261726.11
|
CUX2
|
cut like homeobox 2 |
| chr7_+_54542362 | 0.12 |
ENST00000402613.4
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr11_+_13962676 | 0.12 |
ENST00000576479.4
|
SPON1
|
spondin 1 |
| chr8_+_26293112 | 0.12 |
ENST00000523925.5
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr10_+_24449426 | 0.11 |
ENST00000307544.10
|
KIAA1217
|
KIAA1217 |
| chr9_-_23826231 | 0.11 |
ENST00000397312.7
|
ELAVL2
|
ELAV like RNA binding protein 2 |
| chr17_-_41065879 | 0.10 |
ENST00000394015.3
|
KRTAP2-4
|
keratin associated protein 2-4 |
| chr10_-_13707536 | 0.10 |
ENST00000632570.1
ENST00000477221.2 |
FRMD4A
|
FERM domain containing 4A |
| chrX_-_131289266 | 0.10 |
ENST00000370910.5
ENST00000370901.4 ENST00000361420.8 ENST00000370903.8 |
IGSF1
|
immunoglobulin superfamily member 1 |
| chr6_-_111606260 | 0.10 |
ENST00000340026.10
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr18_+_31591869 | 0.10 |
ENST00000237014.8
|
TTR
|
transthyretin |
| chr10_-_60572599 | 0.09 |
ENST00000503366.5
|
ANK3
|
ankyrin 3 |
| chr14_+_22202561 | 0.08 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr2_-_165203870 | 0.08 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr7_+_90709231 | 0.08 |
ENST00000446790.5
ENST00000265741.7 |
CDK14
|
cyclin dependent kinase 14 |
| chrX_+_106920393 | 0.08 |
ENST00000336803.2
|
CLDN2
|
claudin 2 |
| chr4_-_103077282 | 0.08 |
ENST00000503230.5
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9 member B2 |
| chr1_-_114153863 | 0.07 |
ENST00000610222.3
ENST00000369547.6 ENST00000641643.2 |
SYT6
|
synaptotagmin 6 |
| chr2_-_163735989 | 0.07 |
ENST00000333129.4
ENST00000409634.5 |
FIGN
|
fidgetin, microtubule severing factor |
| chr5_+_141484997 | 0.07 |
ENST00000617094.1
ENST00000306593.2 ENST00000610539.1 ENST00000618371.4 |
PCDHGC4
|
protocadherin gamma subfamily C, 4 |
| chr7_+_90709530 | 0.07 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14 |
| chr11_+_59436469 | 0.07 |
ENST00000641045.1
|
OR5A1
|
olfactory receptor family 5 subfamily A member 1 |
| chr11_+_46701010 | 0.07 |
ENST00000311764.3
|
ZNF408
|
zinc finger protein 408 |
| chr7_+_33904911 | 0.06 |
ENST00000297161.6
|
BMPER
|
BMP binding endothelial regulator |
| chr16_+_70173783 | 0.06 |
ENST00000541793.7
ENST00000314151.12 ENST00000565806.5 ENST00000569347.6 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18 member C |
| chr7_+_90709816 | 0.06 |
ENST00000436577.3
|
CDK14
|
cyclin dependent kinase 14 |
| chr1_+_150364621 | 0.06 |
ENST00000401000.8
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr16_+_69950907 | 0.06 |
ENST00000393701.6
ENST00000568461.5 |
CLEC18A
|
C-type lectin domain family 18 member A |
| chr17_+_17782108 | 0.06 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr9_+_133636355 | 0.06 |
ENST00000393056.8
|
DBH
|
dopamine beta-hydroxylase |
| chr11_-_16356538 | 0.06 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr3_+_130850585 | 0.06 |
ENST00000505330.5
ENST00000504381.5 ENST00000507488.6 |
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr7_+_54542393 | 0.05 |
ENST00000404951.5
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr16_+_69951180 | 0.05 |
ENST00000288040.10
ENST00000449317.6 |
CLEC18A
|
C-type lectin domain family 18 member A |
| chr13_+_75804270 | 0.05 |
ENST00000447038.5
|
LMO7
|
LIM domain 7 |
| chr8_-_102412686 | 0.05 |
ENST00000220959.8
ENST00000520539.6 |
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr7_-_78771108 | 0.05 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr7_+_54542300 | 0.05 |
ENST00000302287.7
ENST00000407838.7 |
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr6_-_138499487 | 0.05 |
ENST00000343505.9
|
NHSL1
|
NHS like 1 |
| chr8_-_42501224 | 0.05 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr11_-_36598221 | 0.05 |
ENST00000311485.8
ENST00000527033.5 ENST00000532616.1 ENST00000618712.4 |
RAG2
|
recombination activating 2 |
| chr18_+_78979811 | 0.05 |
ENST00000537592.7
|
SALL3
|
spalt like transcription factor 3 |
| chr10_+_84194527 | 0.04 |
ENST00000623527.4
|
CDHR1
|
cadherin related family member 1 |
| chr8_+_38974212 | 0.04 |
ENST00000302495.5
|
HTRA4
|
HtrA serine peptidase 4 |
| chr7_+_148590760 | 0.04 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr1_+_150364136 | 0.04 |
ENST00000369068.5
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr1_-_94925759 | 0.04 |
ENST00000415017.1
ENST00000545882.5 |
CNN3
|
calponin 3 |
| chr3_+_32817990 | 0.04 |
ENST00000383763.6
|
TRIM71
|
tripartite motif containing 71 |
| chr7_+_151048526 | 0.04 |
ENST00000349064.10
|
ASIC3
|
acid sensing ion channel subunit 3 |
| chr17_-_48968587 | 0.04 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr5_-_157059109 | 0.03 |
ENST00000523175.6
ENST00000522693.5 |
HAVCR1
|
hepatitis A virus cellular receptor 1 |
| chr5_-_131165272 | 0.03 |
ENST00000675491.1
ENST00000506908.2 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr2_+_170178136 | 0.03 |
ENST00000409044.7
ENST00000408978.9 |
MYO3B
|
myosin IIIB |
| chr10_+_72692125 | 0.03 |
ENST00000373053.7
ENST00000357157.10 |
MCU
|
mitochondrial calcium uniporter |
| chr12_-_102197827 | 0.03 |
ENST00000329406.5
|
PMCH
|
pro-melanin concentrating hormone |
| chr15_+_33968484 | 0.03 |
ENST00000383263.7
|
CHRM5
|
cholinergic receptor muscarinic 5 |
| chr16_-_67483541 | 0.03 |
ENST00000290953.3
|
AGRP
|
agouti related neuropeptide |
| chr11_-_3837858 | 0.03 |
ENST00000396979.1
|
RHOG
|
ras homolog family member G |
| chr11_+_22338333 | 0.03 |
ENST00000263160.4
|
SLC17A6
|
solute carrier family 17 member 6 |
| chr9_-_146140 | 0.03 |
ENST00000475990.5
|
CBWD1
|
COBW domain containing 1 |
| chr4_-_76007501 | 0.02 |
ENST00000264888.6
|
CXCL9
|
C-X-C motif chemokine ligand 9 |
| chr1_+_186296267 | 0.02 |
ENST00000533951.5
ENST00000367482.8 ENST00000635041.1 ENST00000367483.8 ENST00000367485.4 ENST00000445192.7 |
PRG4
|
proteoglycan 4 |
| chr6_-_111605859 | 0.02 |
ENST00000651359.1
ENST00000650859.1 ENST00000359831.8 ENST00000368761.11 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr5_-_83673544 | 0.02 |
ENST00000503117.1
ENST00000510978.5 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chrX_+_131058340 | 0.02 |
ENST00000276211.10
ENST00000370922.5 |
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr4_+_70592295 | 0.02 |
ENST00000449493.2
|
AMBN
|
ameloblastin |
| chr2_+_44941695 | 0.02 |
ENST00000260653.5
|
SIX3
|
SIX homeobox 3 |
| chr2_-_88861258 | 0.01 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr2_-_164842140 | 0.01 |
ENST00000496396.1
ENST00000629362.2 ENST00000445474.2 ENST00000483743.6 |
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr8_-_33567118 | 0.01 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122 |
| chr18_-_76495191 | 0.01 |
ENST00000443185.7
|
ZNF516
|
zinc finger protein 516 |
| chr13_-_26221703 | 0.01 |
ENST00000381570.7
ENST00000346166.7 |
RNF6
|
ring finger protein 6 |
| chr20_+_43507668 | 0.01 |
ENST00000434666.5
ENST00000649084.1 ENST00000418998.7 ENST00000427442.8 ENST00000649917.1 |
L3MBTL1
|
L3MBTL histone methyl-lysine binding protein 1 |
| chr1_+_248508073 | 0.01 |
ENST00000641804.1
|
OR2G6
|
olfactory receptor family 2 subfamily G member 6 |
| chr1_+_78004930 | 0.01 |
ENST00000370763.6
|
DNAJB4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr22_-_40533808 | 0.01 |
ENST00000422851.1
ENST00000651694.1 ENST00000652095.2 |
MRTFA
|
myocardin related transcription factor A |
| chr4_+_70721953 | 0.01 |
ENST00000381006.8
ENST00000226328.8 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr2_+_158968608 | 0.01 |
ENST00000263635.8
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr17_-_10469558 | 0.01 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4 |
| chr10_+_132537814 | 0.00 |
ENST00000368593.7
|
INPP5A
|
inositol polyphosphate-5-phosphatase A |
| chr12_+_55549602 | 0.00 |
ENST00000641569.1
ENST00000641851.1 |
OR6C4
|
olfactory receptor family 6 subfamily C member 4 |
| chr1_+_42825548 | 0.00 |
ENST00000372514.7
|
ERMAP
|
erythroblast membrane associated protein (Scianna blood group) |
| chr11_+_75815180 | 0.00 |
ENST00000356136.8
|
UVRAG
|
UV radiation resistance associated |
| chr2_-_223602284 | 0.00 |
ENST00000421386.1
ENST00000305409.3 ENST00000433889.1 |
SCG2
|
secretogranin II |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.4 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.3 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.2 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.1 | 0.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.0 | 0.1 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.7 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.5 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 1.4 | GO:0002381 | immunoglobulin production involved in immunoglobulin mediated immune response(GO:0002381) |
| 0.0 | 0.2 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.0 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.5 | GO:0030297 | ErbB-2 class receptor binding(GO:0005176) transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |