Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
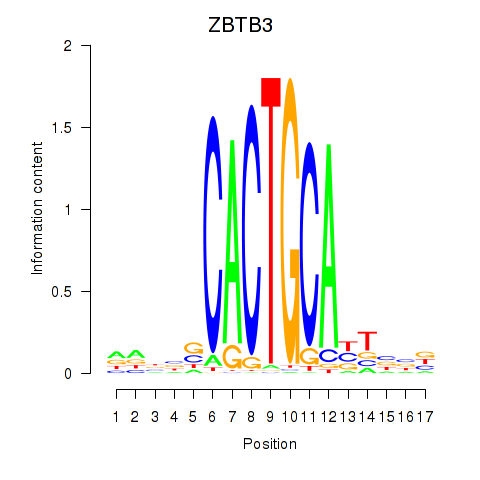
Results for ZBTB3
Z-value: 0.29
Transcription factors associated with ZBTB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB3
|
ENSG00000185670.9 | ZBTB3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB3 | hg38_v1_chr11_-_62754141_62754184 | -0.09 | 6.2e-01 | Click! |
Activity profile of ZBTB3 motif
Sorted Z-values of ZBTB3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_83673544 | 0.49 |
ENST00000503117.1
ENST00000510978.5 |
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr5_-_150290093 | 0.37 |
ENST00000672479.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr20_-_56497608 | 0.28 |
ENST00000617620.1
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr12_-_119803383 | 0.27 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr5_+_76083398 | 0.23 |
ENST00000322285.7
|
SV2C
|
synaptic vesicle glycoprotein 2C |
| chr12_-_119804298 | 0.19 |
ENST00000678652.1
ENST00000678494.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr11_+_123358416 | 0.17 |
ENST00000638157.1
|
GRAMD1B
|
GRAM domain containing 1B |
| chr2_-_2326210 | 0.14 |
ENST00000647755.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr2_-_153478753 | 0.14 |
ENST00000325926.4
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator homolog |
| chr2_-_2326378 | 0.13 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr3_-_52826834 | 0.13 |
ENST00000441637.2
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain 4 |
| chr6_+_29306626 | 0.13 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr20_+_56412249 | 0.12 |
ENST00000679887.1
ENST00000434344.2 |
CASS4
|
Cas scaffold protein family member 4 |
| chr1_-_110606009 | 0.12 |
ENST00000640774.2
ENST00000638616.2 |
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr12_+_48183602 | 0.12 |
ENST00000316554.5
|
CCDC184
|
coiled-coil domain containing 184 |
| chr20_+_56412112 | 0.12 |
ENST00000360314.7
|
CASS4
|
Cas scaffold protein family member 4 |
| chrX_+_153072454 | 0.12 |
ENST00000421798.5
|
PNMA6A
|
PNMA family member 6A |
| chr20_+_56412393 | 0.12 |
ENST00000679529.1
|
CASS4
|
Cas scaffold protein family member 4 |
| chr19_-_6670151 | 0.11 |
ENST00000675206.1
|
TNFSF14
|
TNF superfamily member 14 |
| chr7_+_143935233 | 0.11 |
ENST00000408955.3
|
OR2F2
|
olfactory receptor family 2 subfamily F member 2 |
| chr21_-_46605073 | 0.11 |
ENST00000291700.9
ENST00000367071.4 |
S100B
|
S100 calcium binding protein B |
| chr5_+_170861990 | 0.11 |
ENST00000523189.6
|
RANBP17
|
RAN binding protein 17 |
| chr15_-_74367637 | 0.11 |
ENST00000268053.11
ENST00000416978.1 |
CYP11A1
|
cytochrome P450 family 11 subfamily A member 1 |
| chr19_-_6670117 | 0.11 |
ENST00000245912.7
|
TNFSF14
|
TNF superfamily member 14 |
| chr10_-_116273606 | 0.11 |
ENST00000682743.1
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr17_+_4498866 | 0.10 |
ENST00000329078.8
|
SPNS2
|
sphingolipid transporter 2 |
| chr1_-_110606347 | 0.10 |
ENST00000316361.10
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr2_-_110212519 | 0.09 |
ENST00000611969.5
|
MTLN
|
mitoregulin |
| chr16_-_68372258 | 0.09 |
ENST00000568373.5
ENST00000563226.1 |
SMPD3
|
sphingomyelin phosphodiesterase 3 |
| chr12_+_53097656 | 0.09 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr10_-_75109085 | 0.09 |
ENST00000607131.5
|
DUSP13
|
dual specificity phosphatase 13 |
| chr19_+_35030626 | 0.09 |
ENST00000638536.1
|
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr19_+_35030438 | 0.09 |
ENST00000415950.5
ENST00000262631.11 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr12_+_41188301 | 0.08 |
ENST00000402685.7
|
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr16_-_19884828 | 0.08 |
ENST00000300571.7
ENST00000570142.5 ENST00000562469.5 |
GPRC5B
|
G protein-coupled receptor class C group 5 member B |
| chr10_+_68560317 | 0.08 |
ENST00000373644.5
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr10_-_75109172 | 0.08 |
ENST00000372700.7
ENST00000473072.2 ENST00000491677.6 ENST00000372702.7 |
DUSP13
|
dual specificity phosphatase 13 |
| chr18_+_12407896 | 0.08 |
ENST00000590956.5
ENST00000336990.8 ENST00000440960.6 ENST00000588729.5 |
PRELID3A
|
PRELI domain containing 3A |
| chr2_-_151971750 | 0.07 |
ENST00000636598.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr4_-_5893075 | 0.07 |
ENST00000324989.12
|
CRMP1
|
collapsin response mediator protein 1 |
| chr12_+_54854505 | 0.06 |
ENST00000308796.11
ENST00000619042.1 |
MUCL1
|
mucin like 1 |
| chr1_+_160190567 | 0.06 |
ENST00000368078.8
|
CASQ1
|
calsequestrin 1 |
| chr7_+_100483919 | 0.06 |
ENST00000300179.7
|
NYAP1
|
neuronal tyrosine phosphorylated phosphoinositide-3-kinase adaptor 1 |
| chr10_-_70146602 | 0.06 |
ENST00000335494.5
ENST00000287078.7 |
TYSND1
|
trypsin like peroxisomal matrix peptidase 1 |
| chr6_+_44158807 | 0.06 |
ENST00000532171.5
ENST00000398776.2 |
CAPN11
|
calpain 11 |
| chr11_-_14892217 | 0.05 |
ENST00000334636.10
|
CYP2R1
|
cytochrome P450 family 2 subfamily R member 1 |
| chr16_-_58734299 | 0.05 |
ENST00000245206.10
ENST00000434819.2 |
GOT2
|
glutamic-oxaloacetic transaminase 2 |
| chr15_+_32030481 | 0.05 |
ENST00000454250.7
ENST00000675428.1 ENST00000637552.1 |
CHRNA7
|
cholinergic receptor nicotinic alpha 7 subunit |
| chr4_-_73069696 | 0.05 |
ENST00000507544.3
ENST00000295890.8 |
COX18
|
cytochrome c oxidase assembly factor COX18 |
| chr3_-_39281261 | 0.05 |
ENST00000541347.5
ENST00000412814.1 |
CX3CR1
|
C-X3-C motif chemokine receptor 1 |
| chr11_-_130916437 | 0.05 |
ENST00000533214.1
ENST00000528555.5 ENST00000530356.5 ENST00000265909.9 |
SNX19
|
sorting nexin 19 |
| chr1_+_159171607 | 0.04 |
ENST00000368124.8
ENST00000368125.9 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr7_+_16753731 | 0.04 |
ENST00000262067.5
|
TSPAN13
|
tetraspanin 13 |
| chr16_+_53130921 | 0.04 |
ENST00000564845.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr2_-_2326161 | 0.04 |
ENST00000649810.1
ENST00000648318.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr15_+_69160620 | 0.04 |
ENST00000261858.7
|
GLCE
|
glucuronic acid epimerase |
| chr1_-_155300979 | 0.04 |
ENST00000392414.7
|
PKLR
|
pyruvate kinase L/R |
| chr15_+_32030506 | 0.04 |
ENST00000306901.9
ENST00000636440.1 |
CHRNA7
|
cholinergic receptor nicotinic alpha 7 subunit |
| chr1_+_171257930 | 0.04 |
ENST00000354841.4
|
FMO1
|
flavin containing dimethylaniline monoxygenase 1 |
| chr12_-_43552196 | 0.04 |
ENST00000389420.8
|
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif 20 |
| chr17_-_10421944 | 0.03 |
ENST00000403437.2
|
MYH8
|
myosin heavy chain 8 |
| chr10_+_97572493 | 0.03 |
ENST00000307518.9
ENST00000298808.9 |
ANKRD2
|
ankyrin repeat domain 2 |
| chr13_-_48037934 | 0.03 |
ENST00000646804.1
ENST00000643246.1 |
SUCLA2
|
succinate-CoA ligase ADP-forming subunit beta |
| chr19_+_52345406 | 0.03 |
ENST00000327920.12
|
ZNF610
|
zinc finger protein 610 |
| chr14_-_91946989 | 0.03 |
ENST00000556154.5
|
FBLN5
|
fibulin 5 |
| chrX_+_70234655 | 0.03 |
ENST00000374521.4
|
AWAT1
|
acyl-CoA wax alcohol acyltransferase 1 |
| chr10_+_49609095 | 0.02 |
ENST00000339797.5
|
CHAT
|
choline O-acetyltransferase |
| chrX_+_71144818 | 0.02 |
ENST00000536169.5
ENST00000358741.4 ENST00000395855.6 ENST00000374051.7 |
NLGN3
|
neuroligin 3 |
| chrX_+_135032346 | 0.02 |
ENST00000257013.9
|
RTL8C
|
retrotransposon Gag like 8C |
| chr7_+_134891566 | 0.02 |
ENST00000424922.5
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr5_+_75512058 | 0.02 |
ENST00000514296.5
|
POLK
|
DNA polymerase kappa |
| chr6_-_31972123 | 0.02 |
ENST00000337523.10
|
DXO
|
decapping exoribonuclease |
| chr1_+_171781635 | 0.02 |
ENST00000361735.4
ENST00000362019.7 ENST00000367737.9 |
EEF1AKNMT
|
eEF1A lysine and N-terminal methyltransferase |
| chr3_+_178558700 | 0.02 |
ENST00000432997.5
ENST00000455865.5 |
KCNMB2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr4_-_122621011 | 0.02 |
ENST00000611104.2
ENST00000648588.1 |
IL21
|
interleukin 21 |
| chr19_-_55038256 | 0.02 |
ENST00000417454.5
ENST00000310373.7 ENST00000333884.2 |
GP6
|
glycoprotein VI platelet |
| chr17_+_7627963 | 0.02 |
ENST00000575729.5
ENST00000340624.9 |
SHBG
|
sex hormone binding globulin |
| chr20_+_63658286 | 0.02 |
ENST00000360203.11
ENST00000508582.7 ENST00000318100.9 ENST00000356810.5 |
RTEL1
|
regulator of telomere elongation helicase 1 |
| chr1_-_156053780 | 0.02 |
ENST00000368309.4
|
UBQLN4
|
ubiquilin 4 |
| chr9_+_71911468 | 0.02 |
ENST00000377031.7
|
C9orf85
|
chromosome 9 open reading frame 85 |
| chr19_+_52274443 | 0.02 |
ENST00000600821.5
ENST00000595149.5 ENST00000595000.5 ENST00000593612.1 |
ZNF766
|
zinc finger protein 766 |
| chr5_-_170297746 | 0.02 |
ENST00000046794.10
|
LCP2
|
lymphocyte cytosolic protein 2 |
| chr10_+_44911316 | 0.02 |
ENST00000544540.5
ENST00000389583.5 |
TMEM72
|
transmembrane protein 72 |
| chr14_+_60249191 | 0.02 |
ENST00000395076.9
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent 1A |
| chr6_+_122779707 | 0.02 |
ENST00000368444.8
ENST00000356535.4 |
FABP7
|
fatty acid binding protein 7 |
| chr6_+_30882914 | 0.02 |
ENST00000509639.5
ENST00000412274.6 ENST00000507901.5 ENST00000507046.5 ENST00000437124.6 ENST00000454612.6 ENST00000396342.6 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_+_42169332 | 0.01 |
ENST00000402711.6
ENST00000318522.10 |
EML4
|
EMAP like 4 |
| chr5_+_77086682 | 0.01 |
ENST00000643365.1
ENST00000645183.1 ENST00000645374.1 ENST00000647364.1 ENST00000643848.1 ENST00000643603.1 ENST00000645459.1 ENST00000643269.1 ENST00000503969.6 ENST00000646262.1 |
ZBED3-AS1
ENSG00000284762.1
|
ZBED3 antisense RNA 1 phosphodiesterase 8B |
| chr1_-_204494752 | 0.01 |
ENST00000684373.1
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr10_-_103396441 | 0.01 |
ENST00000309579.7
ENST00000337003.4 ENST00000369815.6 ENST00000369825.6 |
ATP5MK
|
ATP synthase membrane subunit k |
| chr6_+_42173358 | 0.01 |
ENST00000372958.2
|
GUCA1A
|
guanylate cyclase activator 1A |
| chr17_-_58517835 | 0.01 |
ENST00000579921.1
ENST00000579925.5 ENST00000323456.9 |
MTMR4
|
myotubularin related protein 4 |
| chr19_-_39435910 | 0.01 |
ENST00000599539.5
|
RPS16
|
ribosomal protein S16 |
| chr6_-_31972290 | 0.01 |
ENST00000375349.7
|
DXO
|
decapping exoribonuclease |
| chr16_+_30023198 | 0.01 |
ENST00000681219.1
ENST00000300575.6 |
C16orf92
|
chromosome 16 open reading frame 92 |
| chr3_+_98531965 | 0.01 |
ENST00000284311.5
|
GPR15
|
G protein-coupled receptor 15 |
| chr1_+_209827964 | 0.01 |
ENST00000491415.7
|
UTP25
|
UTP25 small subunit processor component |
| chr4_+_158315309 | 0.01 |
ENST00000460056.6
|
RXFP1
|
relaxin family peptide receptor 1 |
| chr11_-_83071819 | 0.01 |
ENST00000524635.1
ENST00000526205.5 ENST00000533486.5 ENST00000533276.6 ENST00000527633.6 |
RAB30
|
RAB30, member RAS oncogene family |
| chr2_-_74343397 | 0.01 |
ENST00000394019.6
ENST00000377634.8 ENST00000436454.1 |
SLC4A5
|
solute carrier family 4 member 5 |
| chr17_-_41467386 | 0.01 |
ENST00000225899.4
|
KRT32
|
keratin 32 |
| chr19_-_2042066 | 0.01 |
ENST00000591588.1
ENST00000591142.5 |
MKNK2
|
MAPK interacting serine/threonine kinase 2 |
| chr19_-_39435627 | 0.01 |
ENST00000599705.1
|
RPS16
|
ribosomal protein S16 |
| chr7_+_134891400 | 0.01 |
ENST00000393118.6
|
CALD1
|
caldesmon 1 |
| chr6_+_138773747 | 0.00 |
ENST00000617445.5
|
CCDC28A
|
coiled-coil domain containing 28A |
| chr2_+_88524640 | 0.00 |
ENST00000303254.4
|
TEX37
|
testis expressed 37 |
| chr8_+_38404363 | 0.00 |
ENST00000527175.1
|
LETM2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chr2_+_42169434 | 0.00 |
ENST00000401738.3
|
EML4
|
EMAP like 4 |
| chr19_-_39435936 | 0.00 |
ENST00000339471.8
ENST00000601655.5 ENST00000251453.8 |
RPS16
|
ribosomal protein S16 |
| chr9_+_71911615 | 0.00 |
ENST00000334731.7
ENST00000486911.2 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr4_-_15938740 | 0.00 |
ENST00000382333.2
|
FGFBP1
|
fibroblast growth factor binding protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.2 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.1 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 | 0.4 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0006532 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.0 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.0 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |