Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
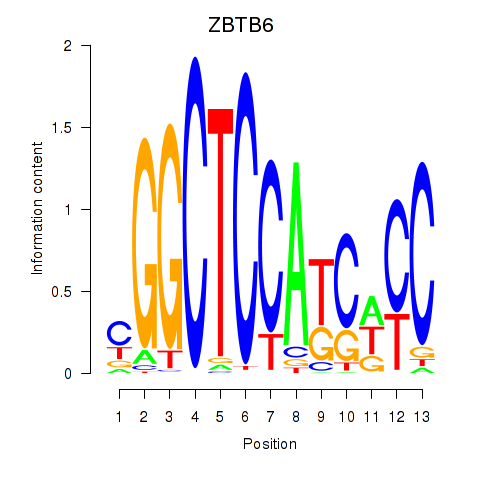
Results for ZBTB6
Z-value: 1.02
Transcription factors associated with ZBTB6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB6
|
ENSG00000186130.5 | ZBTB6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB6 | hg38_v1_chr9_-_122913299_122913333 | -0.12 | 5.3e-01 | Click! |
Activity profile of ZBTB6 motif
Sorted Z-values of ZBTB6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_42496186 | 2.95 |
ENST00000398352.3
ENST00000291536.8 |
RSPH1
|
radial spoke head component 1 |
| chr16_-_52547113 | 2.68 |
ENST00000219746.14
|
TOX3
|
TOX high mobility group box family member 3 |
| chr21_+_34668986 | 2.39 |
ENST00000349499.3
|
CLIC6
|
chloride intracellular channel 6 |
| chr14_-_106622837 | 2.37 |
ENST00000390628.3
|
IGHV1-58
|
immunoglobulin heavy variable 1-58 |
| chr13_+_34942263 | 2.10 |
ENST00000379939.7
ENST00000400445.7 |
NBEA
|
neurobeachin |
| chr9_+_36136416 | 1.65 |
ENST00000396613.7
|
GLIPR2
|
GLI pathogenesis related 2 |
| chr3_-_197959977 | 1.61 |
ENST00000265239.11
|
IQCG
|
IQ motif containing G |
| chr10_-_59709842 | 1.49 |
ENST00000395348.8
|
SLC16A9
|
solute carrier family 16 member 9 |
| chr20_-_34872817 | 1.43 |
ENST00000427420.1
ENST00000336431.10 |
GGT7
|
gamma-glutamyltransferase 7 |
| chr15_+_70892809 | 1.39 |
ENST00000260382.10
ENST00000560755.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr20_-_25058115 | 1.37 |
ENST00000323482.9
|
ACSS1
|
acyl-CoA synthetase short chain family member 1 |
| chr20_-_25058168 | 1.35 |
ENST00000432802.6
|
ACSS1
|
acyl-CoA synthetase short chain family member 1 |
| chr11_+_46277648 | 1.35 |
ENST00000621158.5
|
CREB3L1
|
cAMP responsive element binding protein 3 like 1 |
| chr19_-_55166671 | 1.27 |
ENST00000455045.5
|
DNAAF3
|
dynein axonemal assembly factor 3 |
| chr9_+_87498491 | 1.25 |
ENST00000622514.4
|
DAPK1
|
death associated protein kinase 1 |
| chr2_-_229714478 | 1.25 |
ENST00000341772.5
|
DNER
|
delta/notch like EGF repeat containing |
| chr5_+_140841183 | 1.24 |
ENST00000378123.4
ENST00000531613.2 |
PCDHA8
|
protocadherin alpha 8 |
| chr19_-_55166632 | 1.18 |
ENST00000532817.5
ENST00000527223.6 ENST00000391720.8 |
DNAAF3
|
dynein axonemal assembly factor 3 |
| chr14_+_20684547 | 1.17 |
ENST00000555835.3
ENST00000397995.2 ENST00000553909.1 |
RNASE4
ENSG00000259171.1
|
ribonuclease A family member 4 novel protein, ANG-RNASE4 readthrough |
| chr19_-_55166565 | 1.15 |
ENST00000526003.5
ENST00000534170.5 ENST00000524407.7 |
DNAAF3
|
dynein axonemal assembly factor 3 |
| chr8_-_144060681 | 1.14 |
ENST00000618853.5
|
OPLAH
|
5-oxoprolinase, ATP-hydrolysing |
| chr19_-_5567984 | 1.08 |
ENST00000448587.5
|
TINCR
|
TINCR ubiquitin domain containing |
| chr6_-_31878967 | 1.01 |
ENST00000414427.1
ENST00000229729.11 ENST00000375562.8 |
SLC44A4
|
solute carrier family 44 member 4 |
| chr8_-_40897814 | 1.01 |
ENST00000297737.11
ENST00000315769.11 |
ZMAT4
|
zinc finger matrin-type 4 |
| chr9_+_68705230 | 1.00 |
ENST00000265382.8
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chr3_-_133895577 | 0.99 |
ENST00000543906.5
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr4_-_7042931 | 0.98 |
ENST00000310085.6
|
CCDC96
|
coiled-coil domain containing 96 |
| chr3_-_49132994 | 0.98 |
ENST00000305544.9
ENST00000494831.1 ENST00000418109.5 |
LAMB2
|
laminin subunit beta 2 |
| chr9_+_68705414 | 0.95 |
ENST00000541509.5
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chr19_+_53554819 | 0.94 |
ENST00000505949.5
ENST00000513265.5 ENST00000648511.1 |
ZNF331
|
zinc finger protein 331 |
| chr6_-_6006878 | 0.91 |
ENST00000244766.7
|
NRN1
|
neuritin 1 |
| chr16_+_50742059 | 0.91 |
ENST00000311559.13
ENST00000564326.5 ENST00000566206.5 ENST00000427738.8 |
CYLD
|
CYLD lysine 63 deubiquitinase |
| chr6_-_6007511 | 0.90 |
ENST00000616243.1
|
NRN1
|
neuritin 1 |
| chr14_-_89619118 | 0.89 |
ENST00000345097.8
ENST00000555855.5 ENST00000555353.5 |
FOXN3
|
forkhead box N3 |
| chr10_-_97334698 | 0.88 |
ENST00000371019.4
|
FRAT2
|
FRAT regulator of WNT signaling pathway 2 |
| chr3_+_181711915 | 0.88 |
ENST00000325404.3
|
SOX2
|
SRY-box transcription factor 2 |
| chr16_-_67416420 | 0.88 |
ENST00000348579.6
ENST00000565726.3 |
ZDHHC1
|
zinc finger DHHC-type containing 1 |
| chr9_-_114387973 | 0.84 |
ENST00000374088.8
|
AKNA
|
AT-hook transcription factor |
| chr3_-_133895453 | 0.84 |
ENST00000486858.5
ENST00000477759.5 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr6_-_154510114 | 0.83 |
ENST00000673182.1
|
ENSG00000288520.1
|
novel protein |
| chr11_+_77066948 | 0.83 |
ENST00000527066.5
ENST00000648180.1 ENST00000529629.5 |
CAPN5
|
calpain 5 |
| chr19_+_53554499 | 0.83 |
ENST00000512387.6
ENST00000511567.5 |
ZNF331
|
zinc finger protein 331 |
| chr3_-_133895867 | 0.83 |
ENST00000285208.9
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr6_+_156777882 | 0.82 |
ENST00000350026.10
ENST00000647938.1 ENST00000674298.1 |
ARID1B
|
AT-rich interaction domain 1B |
| chr7_+_76201863 | 0.81 |
ENST00000611745.2
|
SRRM3
|
serine/arginine repetitive matrix 3 |
| chr22_+_31122923 | 0.80 |
ENST00000620191.4
ENST00000412277.6 ENST00000412985.5 ENST00000331075.10 ENST00000420017.5 ENST00000400294.6 ENST00000405300.5 ENST00000404390.7 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr19_-_7926106 | 0.78 |
ENST00000318978.6
|
CTXN1
|
cortexin 1 |
| chr19_+_50649445 | 0.77 |
ENST00000425202.6
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chrX_+_49879475 | 0.76 |
ENST00000621775.2
|
USP27X
|
ubiquitin specific peptidase 27 X-linked |
| chr20_-_63831214 | 0.76 |
ENST00000302995.2
ENST00000245663.9 |
ZBTB46
|
zinc finger and BTB domain containing 46 |
| chr5_+_102755269 | 0.75 |
ENST00000304400.12
ENST00000455264.7 ENST00000684529.1 ENST00000438793.8 ENST00000682882.1 ENST00000682972.1 ENST00000348126.7 ENST00000512073.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr12_+_6821646 | 0.75 |
ENST00000428545.6
|
GPR162
|
G protein-coupled receptor 162 |
| chr10_+_125896549 | 0.74 |
ENST00000368693.6
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr22_-_31345770 | 0.74 |
ENST00000215919.3
|
PATZ1
|
POZ/BTB and AT hook containing zinc finger 1 |
| chr19_+_18001117 | 0.72 |
ENST00000379656.7
|
ARRDC2
|
arrestin domain containing 2 |
| chr6_+_71288803 | 0.71 |
ENST00000370435.5
|
OGFRL1
|
opioid growth factor receptor like 1 |
| chr6_+_29723421 | 0.71 |
ENST00000259951.12
ENST00000434407.6 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr3_+_48466222 | 0.71 |
ENST00000625293.3
ENST00000492235.2 ENST00000635452.2 ENST00000456089.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr9_+_87497675 | 0.70 |
ENST00000472284.5
ENST00000469640.6 |
DAPK1
|
death associated protein kinase 1 |
| chr2_+_119544420 | 0.70 |
ENST00000413369.8
|
CFAP221
|
cilia and flagella associated protein 221 |
| chr19_-_49072699 | 0.70 |
ENST00000221444.2
|
KCNA7
|
potassium voltage-gated channel subfamily A member 7 |
| chr12_+_50057548 | 0.70 |
ENST00000228468.8
ENST00000447966.7 |
ASIC1
|
acid sensing ion channel subunit 1 |
| chr19_+_51311638 | 0.69 |
ENST00000270642.9
|
IGLON5
|
IgLON family member 5 |
| chr11_+_6239068 | 0.69 |
ENST00000379936.3
|
CNGA4
|
cyclic nucleotide gated channel subunit alpha 4 |
| chr8_-_73746830 | 0.69 |
ENST00000524300.6
ENST00000523558.5 ENST00000521210.5 ENST00000355780.9 ENST00000524104.5 ENST00000521736.5 ENST00000521447.5 ENST00000517542.5 ENST00000521451.5 ENST00000521419.5 ENST00000518502.5 |
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr22_-_38088915 | 0.68 |
ENST00000428572.1
|
BAIAP2L2
|
BAR/IMD domain containing adaptor protein 2 like 2 |
| chr10_+_112376193 | 0.68 |
ENST00000433418.6
ENST00000356116.6 ENST00000354273.5 |
ACSL5
|
acyl-CoA synthetase long chain family member 5 |
| chr14_+_73537346 | 0.67 |
ENST00000557556.1
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr14_-_60724300 | 0.67 |
ENST00000556952.3
ENST00000216513.5 |
SIX4
|
SIX homeobox 4 |
| chr4_+_663696 | 0.67 |
ENST00000471824.6
|
PDE6B
|
phosphodiesterase 6B |
| chr17_+_7705193 | 0.66 |
ENST00000226091.3
|
EFNB3
|
ephrin B3 |
| chr19_+_12995554 | 0.64 |
ENST00000397661.6
|
NFIX
|
nuclear factor I X |
| chr22_+_22704265 | 0.64 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 |
| chr15_-_74906613 | 0.64 |
ENST00000565772.5
|
FAM219B
|
family with sequence similarity 219 member B |
| chr7_+_26152188 | 0.63 |
ENST00000056233.4
|
NFE2L3
|
nuclear factor, erythroid 2 like 3 |
| chr16_+_4734519 | 0.63 |
ENST00000299320.10
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr15_+_43517590 | 0.63 |
ENST00000300231.6
|
MAP1A
|
microtubule associated protein 1A |
| chr17_-_42194459 | 0.62 |
ENST00000593209.5
ENST00000588352.5 ENST00000587427.6 ENST00000414034.7 ENST00000590249.5 ENST00000428494.6 |
GHDC
|
GH3 domain containing |
| chr9_+_34958254 | 0.61 |
ENST00000242315.3
|
PHF24
|
PHD finger protein 24 |
| chr5_+_102754631 | 0.61 |
ENST00000510208.2
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr16_+_86566821 | 0.61 |
ENST00000649859.1
|
FOXC2
|
forkhead box C2 |
| chr6_+_29723340 | 0.61 |
ENST00000334668.8
|
HLA-F
|
major histocompatibility complex, class I, F |
| chr19_+_44809053 | 0.61 |
ENST00000611077.5
|
BCAM
|
basal cell adhesion molecule (Lutheran blood group) |
| chr7_+_94908230 | 0.61 |
ENST00000433881.5
|
PPP1R9A
|
protein phosphatase 1 regulatory subunit 9A |
| chr15_-_43266857 | 0.59 |
ENST00000349114.8
ENST00000220420.10 |
TGM5
|
transglutaminase 5 |
| chr1_+_87331668 | 0.59 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr1_-_151826085 | 0.59 |
ENST00000356728.11
|
RORC
|
RAR related orphan receptor C |
| chr11_+_68460712 | 0.58 |
ENST00000528635.5
ENST00000533127.5 ENST00000529907.5 ENST00000529344.5 ENST00000534534.5 ENST00000524845.5 ENST00000393800.7 ENST00000265637.8 ENST00000524904.5 ENST00000393801.7 ENST00000265636.9 ENST00000529710.5 |
PPP6R3
|
protein phosphatase 6 regulatory subunit 3 |
| chr8_-_12194067 | 0.57 |
ENST00000524571.6
ENST00000533852.6 ENST00000533513.1 ENST00000448228.6 ENST00000534520.5 |
FAM86B1
|
family with sequence similarity 86 member B1 |
| chr13_+_98143410 | 0.57 |
ENST00000596580.2
ENST00000376581.9 |
FARP1
|
FERM, ARH/RhoGEF and pleckstrin domain protein 1 |
| chr3_-_52830664 | 0.57 |
ENST00000266041.9
ENST00000406595.5 ENST00000485816.5 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain 4 |
| chr19_-_51026593 | 0.57 |
ENST00000600362.5
ENST00000453757.8 ENST00000601671.1 |
KLK11
|
kallikrein related peptidase 11 |
| chr12_+_6821797 | 0.56 |
ENST00000311268.8
ENST00000382315.7 |
GPR162
|
G protein-coupled receptor 162 |
| chr1_-_91886144 | 0.56 |
ENST00000212355.9
|
TGFBR3
|
transforming growth factor beta receptor 3 |
| chr12_-_6700788 | 0.56 |
ENST00000320591.9
ENST00000534837.6 |
PIANP
|
PILR alpha associated neural protein |
| chr16_+_50742037 | 0.56 |
ENST00000569418.5
|
CYLD
|
CYLD lysine 63 deubiquitinase |
| chr19_+_43716095 | 0.56 |
ENST00000596627.1
|
IRGC
|
immunity related GTPase cinema |
| chr11_+_77066985 | 0.55 |
ENST00000456580.6
|
CAPN5
|
calpain 5 |
| chr1_-_223364059 | 0.55 |
ENST00000343846.7
ENST00000484758.6 ENST00000344029.6 ENST00000366878.9 ENST00000494793.6 ENST00000681285.1 ENST00000680429.1 ENST00000681669.1 ENST00000681305.1 |
SUSD4
|
sushi domain containing 4 |
| chr21_-_46323806 | 0.55 |
ENST00000397680.5
ENST00000445935.5 ENST00000291691.12 ENST00000397682.7 |
C21orf58
|
chromosome 21 open reading frame 58 |
| chr19_-_5567831 | 0.55 |
ENST00000587632.1
ENST00000646160.1 |
TINCR
|
TINCR ubiquitin domain containing |
| chrX_-_54183207 | 0.55 |
ENST00000375180.7
ENST00000328235.4 ENST00000477084.1 |
FAM120C
|
family with sequence similarity 120C |
| chr17_+_17303319 | 0.55 |
ENST00000616989.1
ENST00000389022.9 |
NT5M
|
5',3'-nucleotidase, mitochondrial |
| chr1_+_21570303 | 0.54 |
ENST00000374830.2
|
ALPL
|
alkaline phosphatase, biomineralization associated |
| chr19_+_53538415 | 0.54 |
ENST00000648122.1
|
ZNF331
|
zinc finger protein 331 |
| chr19_+_53521245 | 0.54 |
ENST00000649326.1
|
ZNF331
|
zinc finger protein 331 |
| chr20_+_23035312 | 0.54 |
ENST00000255008.5
|
SSTR4
|
somatostatin receptor 4 |
| chr17_-_45262084 | 0.53 |
ENST00000331780.5
|
SPATA32
|
spermatogenesis associated 32 |
| chr12_+_93377883 | 0.53 |
ENST00000337179.9
ENST00000415493.7 |
NUDT4
|
nudix hydrolase 4 |
| chr22_-_31292445 | 0.53 |
ENST00000402249.7
ENST00000215912.10 ENST00000443175.1 ENST00000441972.5 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr7_+_118214633 | 0.53 |
ENST00000477532.5
|
ANKRD7
|
ankyrin repeat domain 7 |
| chr19_+_37078411 | 0.52 |
ENST00000337995.4
ENST00000304239.11 ENST00000589245.5 |
ZNF420
|
zinc finger protein 420 |
| chrX_+_40581035 | 0.52 |
ENST00000447485.6
|
ATP6AP2
|
ATPase H+ transporting accessory protein 2 |
| chr16_+_50742110 | 0.52 |
ENST00000566679.6
ENST00000564634.5 ENST00000398568.6 |
CYLD
|
CYLD lysine 63 deubiquitinase |
| chr20_-_59032292 | 0.52 |
ENST00000395663.1
ENST00000243997.8 ENST00000395659.1 |
ATP5F1E
|
ATP synthase F1 subunit epsilon |
| chr10_+_80132722 | 0.52 |
ENST00000372263.4
|
PLAC9
|
placenta associated 9 |
| chr9_+_127706975 | 0.51 |
ENST00000373295.7
|
CFAP157
|
cilia and flagella associated protein 157 |
| chr4_-_102760914 | 0.51 |
ENST00000505239.1
ENST00000647097.2 |
MANBA
|
mannosidase beta |
| chr19_+_12995467 | 0.51 |
ENST00000592199.6
|
NFIX
|
nuclear factor I X |
| chr11_+_73308237 | 0.50 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17 |
| chr22_-_38844020 | 0.50 |
ENST00000333039.4
|
NPTXR
|
neuronal pentraxin receptor |
| chr3_+_194685874 | 0.50 |
ENST00000329759.6
|
FAM43A
|
family with sequence similarity 43 member A |
| chr7_+_7968787 | 0.50 |
ENST00000223145.10
|
GLCCI1
|
glucocorticoid induced 1 |
| chr1_-_54406385 | 0.50 |
ENST00000610401.5
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr4_-_102760976 | 0.50 |
ENST00000644159.1
ENST00000642252.1 |
MANBA
|
mannosidase beta |
| chr2_+_12716893 | 0.49 |
ENST00000381465.2
ENST00000155926.9 |
TRIB2
|
tribbles pseudokinase 2 |
| chr14_-_25049889 | 0.49 |
ENST00000419632.6
ENST00000396700.5 |
STXBP6
|
syntaxin binding protein 6 |
| chr11_-_72794032 | 0.49 |
ENST00000334805.11
|
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr19_+_7507082 | 0.49 |
ENST00000599312.1
|
ENSG00000267952.1
|
novel transcript |
| chr3_+_152835122 | 0.49 |
ENST00000305097.6
|
P2RY1
|
purinergic receptor P2Y1 |
| chr11_+_47980538 | 0.49 |
ENST00000613246.4
ENST00000418331.7 ENST00000615445.4 ENST00000440289.6 |
PTPRJ
|
protein tyrosine phosphatase receptor type J |
| chr22_-_30574654 | 0.48 |
ENST00000406361.6
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr17_-_11997372 | 0.48 |
ENST00000322748.7
ENST00000454073.7 ENST00000580306.7 ENST00000580903.1 |
ZNF18
|
zinc finger protein 18 |
| chr5_+_51383394 | 0.48 |
ENST00000230658.12
|
ISL1
|
ISL LIM homeobox 1 |
| chr2_+_147844488 | 0.48 |
ENST00000535787.5
|
ACVR2A
|
activin A receptor type 2A |
| chr2_-_218010202 | 0.48 |
ENST00000646520.1
|
TNS1
|
tensin 1 |
| chr6_-_31728877 | 0.48 |
ENST00000437288.5
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr4_-_141132568 | 0.47 |
ENST00000506101.2
|
RNF150
|
ring finger protein 150 |
| chr19_+_51127030 | 0.47 |
ENST00000599948.1
|
SIGLEC9
|
sialic acid binding Ig like lectin 9 |
| chr17_-_41027198 | 0.47 |
ENST00000361883.6
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr11_-_62591554 | 0.47 |
ENST00000494385.1
ENST00000308436.11 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr11_+_61752603 | 0.46 |
ENST00000278836.10
|
MYRF
|
myelin regulatory factor |
| chr4_-_25863537 | 0.46 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr6_-_31729260 | 0.46 |
ENST00000375789.7
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr22_-_31489761 | 0.46 |
ENST00000344710.9
ENST00000330125.10 ENST00000397518.5 |
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr4_-_25862979 | 0.46 |
ENST00000399878.8
|
SEL1L3
|
SEL1L family member 3 |
| chr12_+_64780465 | 0.46 |
ENST00000542120.6
|
TBC1D30
|
TBC1 domain family member 30 |
| chr2_+_147845020 | 0.45 |
ENST00000241416.12
|
ACVR2A
|
activin A receptor type 2A |
| chr6_-_24666854 | 0.45 |
ENST00000378198.9
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr20_+_64255728 | 0.45 |
ENST00000369758.8
ENST00000308824.11 ENST00000609372.1 ENST00000610196.1 ENST00000609764.1 |
PCMTD2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr3_-_51499950 | 0.45 |
ENST00000423656.5
ENST00000504652.5 ENST00000684031.1 |
DCAF1
|
DDB1 and CUL4 associated factor 1 |
| chr2_+_95346649 | 0.45 |
ENST00000468529.1
|
KCNIP3
|
potassium voltage-gated channel interacting protein 3 |
| chr5_+_68215738 | 0.44 |
ENST00000521381.6
ENST00000521657.5 |
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr7_-_101180215 | 0.44 |
ENST00000455377.5
ENST00000443096.1 ENST00000300303.7 |
NAT16
|
N-acetyltransferase 16 (putative) |
| chr15_+_45129933 | 0.44 |
ENST00000321429.8
ENST00000389037.7 ENST00000558322.5 |
DUOX1
|
dual oxidase 1 |
| chr4_+_81030700 | 0.44 |
ENST00000282701.4
|
BMP3
|
bone morphogenetic protein 3 |
| chr20_-_63006961 | 0.43 |
ENST00000612929.2
ENST00000370346.2 |
BHLHE23
|
basic helix-loop-helix family member e23 |
| chr10_-_21174187 | 0.43 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr11_+_45922640 | 0.43 |
ENST00000401752.6
ENST00000325468.9 |
LARGE2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr9_+_87497222 | 0.43 |
ENST00000358077.9
|
DAPK1
|
death associated protein kinase 1 |
| chr6_+_147204405 | 0.43 |
ENST00000546097.5
ENST00000367481.7 |
STXBP5
|
syntaxin binding protein 5 |
| chr1_+_154567820 | 0.43 |
ENST00000637900.1
|
CHRNB2
|
cholinergic receptor nicotinic beta 2 subunit |
| chr1_-_107965009 | 0.43 |
ENST00000527011.5
ENST00000370056.9 |
VAV3
|
vav guanine nucleotide exchange factor 3 |
| chr20_+_2692736 | 0.43 |
ENST00000380648.9
ENST00000497450.5 |
EBF4
|
EBF family member 4 |
| chr3_+_141876124 | 0.42 |
ENST00000475483.5
|
ATP1B3
|
ATPase Na+/K+ transporting subunit beta 3 |
| chr5_-_132556809 | 0.42 |
ENST00000450655.1
|
IL5
|
interleukin 5 |
| chr7_-_102616692 | 0.42 |
ENST00000521076.5
ENST00000462172.5 ENST00000522801.5 ENST00000262940.12 ENST00000449970.6 |
RASA4
|
RAS p21 protein activator 4 |
| chr17_+_59565598 | 0.42 |
ENST00000251241.9
ENST00000425628.7 ENST00000584385.5 ENST00000580030.1 |
DHX40
|
DEAH-box helicase 40 |
| chr3_-_184711947 | 0.42 |
ENST00000317897.5
|
MAGEF1
|
MAGE family member F1 |
| chr3_-_8769602 | 0.42 |
ENST00000316793.8
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr6_-_24666591 | 0.42 |
ENST00000341060.3
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr6_-_37697875 | 0.42 |
ENST00000434837.8
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr22_-_30574572 | 0.42 |
ENST00000402369.5
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr19_-_42242526 | 0.42 |
ENST00000222330.8
ENST00000676537.1 |
GSK3A
|
glycogen synthase kinase 3 alpha |
| chr3_+_129001298 | 0.42 |
ENST00000683648.1
|
EFCC1
|
EF-hand and coiled-coil domain containing 1 |
| chr12_-_51026325 | 0.42 |
ENST00000547198.5
ENST00000643884.1 |
SLC11A2
|
solute carrier family 11 member 2 |
| chr12_-_51026345 | 0.41 |
ENST00000547732.5
ENST00000644495.1 ENST00000262052.9 ENST00000546488.5 ENST00000550714.5 ENST00000548193.5 ENST00000547579.5 ENST00000546743.5 |
SLC11A2
|
solute carrier family 11 member 2 |
| chr14_+_22271921 | 0.41 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr19_+_53520948 | 0.41 |
ENST00000648236.1
ENST00000253144.13 |
ZNF331
|
zinc finger protein 331 |
| chr16_+_30740621 | 0.41 |
ENST00000615541.3
ENST00000483578.1 |
TMEM265
ENSG00000260899.1
|
transmembrane protein 265 novel transcript |
| chr8_-_29263063 | 0.41 |
ENST00000524189.6
|
KIF13B
|
kinesin family member 13B |
| chr1_-_160070150 | 0.40 |
ENST00000644903.1
|
KCNJ10
|
potassium inwardly rectifying channel subfamily J member 10 |
| chr12_-_51026141 | 0.40 |
ENST00000541174.6
|
SLC11A2
|
solute carrier family 11 member 2 |
| chr19_-_14979676 | 0.40 |
ENST00000598504.5
ENST00000597262.1 |
SLC1A6
|
solute carrier family 1 member 6 |
| chr16_-_31010611 | 0.40 |
ENST00000215095.11
|
STX1B
|
syntaxin 1B |
| chr3_+_129001581 | 0.40 |
ENST00000436022.2
|
EFCC1
|
EF-hand and coiled-coil domain containing 1 |
| chr12_-_6700377 | 0.40 |
ENST00000540656.5
|
PIANP
|
PILR alpha associated neural protein |
| chr20_+_62656359 | 0.40 |
ENST00000370507.5
|
SLCO4A1
|
solute carrier organic anion transporter family member 4A1 |
| chr3_-_123027721 | 0.40 |
ENST00000357599.8
ENST00000195173.8 ENST00000648990.1 |
SEMA5B
|
semaphorin 5B |
| chr12_+_459925 | 0.40 |
ENST00000266383.10
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyltransferase 3 |
| chr3_+_112333147 | 0.40 |
ENST00000473539.5
ENST00000315711.12 ENST00000383681.7 |
CD200
|
CD200 molecule |
| chr19_+_40613416 | 0.40 |
ENST00000599724.5
ENST00000597071.5 |
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr17_+_4143168 | 0.39 |
ENST00000577075.6
ENST00000301391.8 ENST00000575251.5 |
CYB5D2
|
cytochrome b5 domain containing 2 |
| chr1_-_236281951 | 0.39 |
ENST00000354619.10
|
ERO1B
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr6_+_30940970 | 0.39 |
ENST00000462446.6
ENST00000304311.3 |
MUCL3
|
mucin like 3 |
| chr8_-_21788853 | 0.39 |
ENST00000524240.6
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr19_-_19733091 | 0.39 |
ENST00000344099.4
|
ZNF14
|
zinc finger protein 14 |
| chr20_-_2840623 | 0.39 |
ENST00000360652.7
ENST00000448755.5 |
PCED1A
|
PC-esterase domain containing 1A |
| chr1_-_160070102 | 0.39 |
ENST00000638728.1
ENST00000637644.1 |
KCNJ10
|
potassium inwardly rectifying channel subfamily J member 10 |
| chr19_-_57814878 | 0.39 |
ENST00000391701.1
|
ZNF552
|
zinc finger protein 552 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.5 | 2.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.3 | 1.4 | GO:0018032 | protein amidation(GO:0018032) |
| 0.3 | 1.0 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.3 | 1.0 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.3 | 1.0 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.2 | 1.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.2 | 0.7 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.2 | 0.6 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.2 | 1.6 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.2 | 0.5 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.2 | 0.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 1.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 0.5 | GO:0060913 | visceral motor neuron differentiation(GO:0021524) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.2 | 0.8 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 2.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.6 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.7 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 1.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.1 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 | 0.4 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.4 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.5 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 4.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.0 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.3 | GO:0019483 | uracil catabolic process(GO:0006212) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.3 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.1 | 0.4 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.5 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.4 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.3 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.4 | GO:2000077 | negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.1 | 0.7 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.1 | 0.8 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.3 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.6 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.4 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 2.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 1.3 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.1 | 0.7 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) floor plate development(GO:0033504) |
| 0.1 | 0.5 | GO:0071543 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.4 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.8 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 | 0.7 | GO:0060405 | regulation of penile erection(GO:0060405) |
| 0.1 | 0.3 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.1 | 1.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.2 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 2.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.6 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.2 | GO:0033168 | conversion of ds siRNA to ss siRNA involved in RNA interference(GO:0033168) conversion of ds siRNA to ss siRNA(GO:0036404) |
| 0.1 | 2.9 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.3 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.1 | 0.2 | GO:0003273 | mitral valve formation(GO:0003192) cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.1 | 1.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 1.0 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.2 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 1.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.2 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.2 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.4 | GO:0014057 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) acetylcholine secretion(GO:0061526) |
| 0.1 | 0.2 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 0.5 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.1 | 0.2 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 0.7 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.1 | 0.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.3 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.1 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.3 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 0.9 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.2 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.5 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 3.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 0.3 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.3 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.6 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.5 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 2.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.5 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.3 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:0036047 | protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) peptidyl-lysine deglutarylation(GO:0061699) |
| 0.0 | 0.5 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.4 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.3 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 1.3 | GO:0097502 | mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 1.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.9 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.2 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.1 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.1 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 2.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.3 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.8 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.1 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.0 | 0.2 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0045113 | integrin biosynthetic process(GO:0045112) regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.4 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.5 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 1.0 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.7 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.7 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.6 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.4 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 2.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.0 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.3 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.3 | GO:0097286 | iron ion import(GO:0097286) |
| 0.0 | 0.2 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:1901253 | mesodermal cell fate determination(GO:0007500) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.0 | GO:0002296 | T-helper cell lineage commitment(GO:0002295) T-helper 1 cell lineage commitment(GO:0002296) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0048541 | mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) |
| 0.0 | 0.6 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.0 | GO:1901389 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.8 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.2 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.0 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.0 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.9 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.2 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.1 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.2 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.3 | 1.7 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 2.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 1.6 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.5 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 1.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.6 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 2.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.4 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 0.3 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 0.6 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.2 | GO:0033167 | ARC complex(GO:0033167) |
| 0.1 | 0.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.2 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 2.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.4 | GO:0098560 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.7 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.3 | GO:0030663 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 2.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.0 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 2.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.7 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.0 | GO:1990917 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 2.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 2.0 | GO:0034705 | potassium channel complex(GO:0034705) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.5 | 1.6 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.3 | 1.4 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.3 | 1.0 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.3 | 2.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.3 | 0.9 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 1.2 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 0.7 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.2 | 0.6 | GO:0005055 | laminin receptor activity(GO:0005055) |
| 0.2 | 0.6 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.2 | 1.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.2 | 1.3 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.2 | 1.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 2.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.4 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.1 | 0.4 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 1.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.6 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.9 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.8 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.5 | GO:0052841 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 2.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.2 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.3 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.8 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 0.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 1.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 2.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.3 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.2 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.2 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.1 | 1.0 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.1 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.2 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.1 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.0 | 0.4 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 2.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 2.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0036054 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 1.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.3 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 1.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.1 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 1.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.0 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.4 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0089720 | death effector domain binding(GO:0035877) caspase binding(GO:0089720) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.4 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.0 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) CTP binding(GO:0002135) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.2 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.0 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.3 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 2.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.9 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.5 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.6 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 5.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |