Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
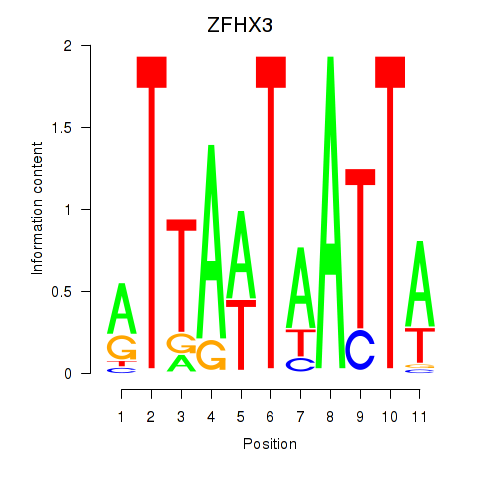
Results for ZFHX3
Z-value: 0.35
Transcription factors associated with ZFHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFHX3
|
ENSG00000140836.17 | ZFHX3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZFHX3 | hg38_v1_chr16_-_73048104_73048136 | 0.37 | 4.6e-02 | Click! |
Activity profile of ZFHX3 motif
Sorted Z-values of ZFHX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZFHX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_735026 | 1.50 |
ENST00000592155.5
ENST00000590161.2 |
PALM
|
paralemmin |
| chr4_-_148444674 | 1.16 |
ENST00000344721.8
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2 |
| chr20_-_7940444 | 0.67 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr5_-_35195236 | 0.58 |
ENST00000509839.5
|
PRLR
|
prolactin receptor |
| chr1_-_247758680 | 0.48 |
ENST00000408896.4
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1 |
| chr15_+_76336755 | 0.43 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr1_-_197067234 | 0.39 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr11_-_119196769 | 0.33 |
ENST00000415318.2
|
CCDC153
|
coiled-coil domain containing 153 |
| chr12_-_7665897 | 0.32 |
ENST00000229304.5
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr16_+_56191476 | 0.31 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1 |
| chr4_-_99435336 | 0.31 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr18_+_58341038 | 0.30 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr3_-_114758940 | 0.30 |
ENST00000464560.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr12_-_130716264 | 0.29 |
ENST00000643940.1
|
RIMBP2
|
RIMS binding protein 2 |
| chr4_-_99435134 | 0.28 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr7_-_100428657 | 0.28 |
ENST00000360951.8
ENST00000398027.6 ENST00000684423.1 ENST00000472716.1 |
ZCWPW1
|
zinc finger CW-type and PWWP domain containing 1 |
| chrX_-_15600953 | 0.28 |
ENST00000679212.1
ENST00000679278.1 ENST00000678046.1 ENST00000252519.8 |
ACE2
|
angiotensin I converting enzyme 2 |
| chr3_+_69866217 | 0.28 |
ENST00000314589.10
|
MITF
|
melanocyte inducing transcription factor |
| chr6_+_25754699 | 0.23 |
ENST00000439485.6
ENST00000377905.9 |
SLC17A4
|
solute carrier family 17 member 4 |
| chr7_+_48171451 | 0.23 |
ENST00000435803.6
|
ABCA13
|
ATP binding cassette subfamily A member 13 |
| chr9_-_121050264 | 0.23 |
ENST00000223642.3
|
C5
|
complement C5 |
| chr2_-_60553618 | 0.23 |
ENST00000643716.1
ENST00000359629.10 ENST00000642384.2 ENST00000335712.11 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr6_-_29045175 | 0.22 |
ENST00000377175.2
|
OR2W1
|
olfactory receptor family 2 subfamily W member 1 |
| chr14_+_39233908 | 0.22 |
ENST00000280082.4
|
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr11_-_61920267 | 0.19 |
ENST00000531922.2
ENST00000301773.9 |
RAB3IL1
|
RAB3A interacting protein like 1 |
| chr19_+_49513353 | 0.19 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr17_+_4788926 | 0.19 |
ENST00000331264.8
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr22_-_30574572 | 0.17 |
ENST00000402369.5
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr17_-_28406160 | 0.17 |
ENST00000618626.1
ENST00000612814.5 |
SLC46A1
|
solute carrier family 46 member 1 |
| chr9_-_123184233 | 0.17 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chrM_+_9207 | 0.15 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr17_+_7407838 | 0.15 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chr12_+_53954870 | 0.15 |
ENST00000243103.4
|
HOXC12
|
homeobox C12 |
| chrX_-_15601077 | 0.14 |
ENST00000680121.1
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr17_+_42900791 | 0.14 |
ENST00000592383.5
ENST00000253801.7 ENST00000585489.1 |
G6PC1
|
glucose-6-phosphatase catalytic subunit 1 |
| chr7_-_7535863 | 0.14 |
ENST00000399429.8
|
COL28A1
|
collagen type XXVIII alpha 1 chain |
| chr4_+_73481737 | 0.14 |
ENST00000226355.5
|
AFM
|
afamin |
| chr11_+_8019193 | 0.13 |
ENST00000534099.5
|
TUB
|
TUB bipartite transcription factor |
| chr2_-_60553409 | 0.12 |
ENST00000358510.6
ENST00000643004.1 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chrY_+_18546691 | 0.12 |
ENST00000309834.8
ENST00000307393.3 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor Y-linked 1 |
| chr1_-_46551647 | 0.12 |
ENST00000481882.7
|
KNCN
|
kinocilin |
| chr18_-_55422306 | 0.12 |
ENST00000566777.5
ENST00000626584.2 |
TCF4
|
transcription factor 4 |
| chr5_+_141245384 | 0.11 |
ENST00000623671.1
ENST00000231173.6 |
PCDHB15
|
protocadherin beta 15 |
| chr18_+_34976928 | 0.11 |
ENST00000591734.5
ENST00000413393.5 ENST00000589180.5 ENST00000587359.5 |
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr4_-_154590735 | 0.11 |
ENST00000403106.8
ENST00000622532.1 ENST00000651975.1 |
FGA
|
fibrinogen alpha chain |
| chr8_-_69833338 | 0.11 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr17_+_17782108 | 0.10 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr6_-_116126120 | 0.10 |
ENST00000452729.1
ENST00000651968.1 ENST00000243222.8 |
COL10A1
|
collagen type X alpha 1 chain |
| chr1_+_76074698 | 0.09 |
ENST00000328299.4
|
ST6GALNAC3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr3_-_157503574 | 0.09 |
ENST00000494677.5
ENST00000468233.5 |
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr10_-_126521439 | 0.09 |
ENST00000284694.11
ENST00000432642.5 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr3_-_157503339 | 0.09 |
ENST00000392833.6
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr2_-_2324642 | 0.09 |
ENST00000650485.1
ENST00000649207.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr2_+_87748087 | 0.09 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2 |
| chr3_-_157503375 | 0.08 |
ENST00000362010.7
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr3_+_77039836 | 0.08 |
ENST00000461745.5
|
ROBO2
|
roundabout guidance receptor 2 |
| chr17_-_69244846 | 0.08 |
ENST00000269081.8
|
ABCA10
|
ATP binding cassette subfamily A member 10 |
| chr7_+_33904911 | 0.08 |
ENST00000297161.6
|
BMPER
|
BMP binding endothelial regulator |
| chr7_+_100428782 | 0.07 |
ENST00000414441.5
|
MEPCE
|
methylphosphate capping enzyme |
| chr6_-_32407123 | 0.07 |
ENST00000374993.4
ENST00000544175.2 ENST00000454136.7 ENST00000446536.2 |
BTNL2
|
butyrophilin like 2 |
| chr2_-_2324323 | 0.06 |
ENST00000648339.1
ENST00000647694.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr6_-_154430495 | 0.06 |
ENST00000424998.3
|
CNKSR3
|
CNKSR family member 3 |
| chr1_-_160579439 | 0.06 |
ENST00000368054.8
ENST00000368048.7 ENST00000311224.8 ENST00000368051.3 ENST00000534968.5 |
CD84
|
CD84 molecule |
| chr12_-_2835018 | 0.05 |
ENST00000337508.9
|
NRIP2
|
nuclear receptor interacting protein 2 |
| chrX_-_123623155 | 0.05 |
ENST00000618150.4
|
THOC2
|
THO complex 2 |
| chr12_-_68159732 | 0.05 |
ENST00000229135.4
|
IFNG
|
interferon gamma |
| chr11_-_102705737 | 0.04 |
ENST00000260229.5
|
MMP27
|
matrix metallopeptidase 27 |
| chr15_-_84654270 | 0.04 |
ENST00000434634.7
|
WDR73
|
WD repeat domain 73 |
| chr6_-_116060859 | 0.04 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr12_+_64497968 | 0.03 |
ENST00000676593.1
ENST00000677093.1 |
TBK1
ENSG00000288665.1
|
TANK binding kinase 1 novel transcript |
| chr20_-_37178966 | 0.03 |
ENST00000422138.1
|
MROH8
|
maestro heat like repeat family member 8 |
| chr1_-_244860376 | 0.03 |
ENST00000638716.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U |
| chr7_-_44541262 | 0.03 |
ENST00000289547.8
ENST00000546276.5 ENST00000423141.1 |
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr1_+_159587817 | 0.03 |
ENST00000255040.3
|
APCS
|
amyloid P component, serum |
| chr2_-_60553558 | 0.03 |
ENST00000642439.1
ENST00000356842.9 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr3_-_165078480 | 0.03 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase |
| chr4_-_52020332 | 0.02 |
ENST00000682860.1
|
LRRC66
|
leucine rich repeat containing 66 |
| chr19_+_53962925 | 0.02 |
ENST00000270458.4
|
CACNG8
|
calcium voltage-gated channel auxiliary subunit gamma 8 |
| chr12_-_55296569 | 0.02 |
ENST00000358433.3
|
OR6C6
|
olfactory receptor family 6 subfamily C member 6 |
| chr12_+_53441724 | 0.02 |
ENST00000551003.5
ENST00000429243.7 ENST00000549068.5 ENST00000549740.5 ENST00000546581.5 ENST00000549581.5 ENST00000547368.5 ENST00000379786.8 ENST00000551945.5 ENST00000547717.1 |
PRR13
ENSG00000257379.1
|
proline rich 13 novel transcript |
| chrM_+_8489 | 0.02 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase membrane subunit 6 |
| chrX_+_85003863 | 0.01 |
ENST00000373173.7
|
APOOL
|
apolipoprotein O like |
| chr14_+_94561435 | 0.01 |
ENST00000557004.6
ENST00000555095.5 ENST00000298841.5 ENST00000554220.5 ENST00000553780.5 |
SERPINA4
SERPINA5
|
serpin family A member 4 serpin family A member 5 |
| chr4_+_99574812 | 0.01 |
ENST00000422897.6
ENST00000265517.10 |
MTTP
|
microsomal triglyceride transfer protein |
| chr2_+_132416795 | 0.01 |
ENST00000329321.4
|
GPR39
|
G protein-coupled receptor 39 |
| chrM_+_10055 | 0.01 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 3 |
| chrX_-_18672101 | 0.01 |
ENST00000379984.4
|
RS1
|
retinoschisin 1 |
| chr20_+_43558968 | 0.01 |
ENST00000647834.1
ENST00000373100.7 ENST00000648083.1 ENST00000648530.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr4_-_53591477 | 0.01 |
ENST00000263925.8
|
LNX1
|
ligand of numb-protein X 1 |
| chr12_-_101407727 | 0.00 |
ENST00000539055.5
ENST00000551688.1 ENST00000551671.5 ENST00000261636.13 |
ARL1
|
ADP ribosylation factor like GTPase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.6 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.4 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.3 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.4 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 | 0.7 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.4 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.1 | 0.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:2000309 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.3 | 1.3 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.4 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |