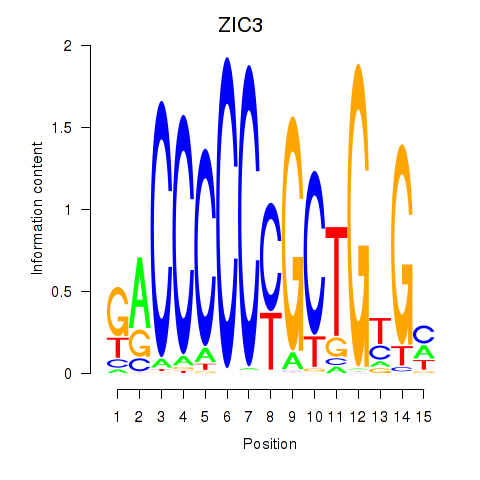
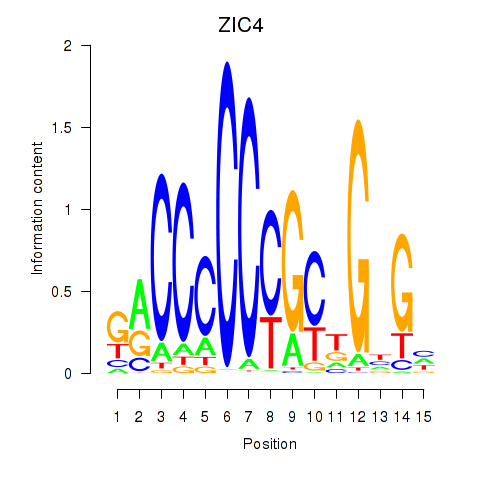
|
chr10_-_13099652
Show fit
|
5.00 |
ENST00000378839.1
|
CCDC3
|
coiled-coil domain containing 3
|
|
chr12_+_49961864
Show fit
|
3.76 |
ENST00000293599.7
|
AQP5
|
aquaporin 5
|
|
chr5_-_180591488
Show fit
|
3.67 |
ENST00000292641.4
|
SCGB3A1
|
secretoglobin family 3A member 1
|
|
chr5_+_77210881
Show fit
|
2.96 |
ENST00000340978.7
ENST00000346042.7
ENST00000342343.8
ENST00000333194.8
|
PDE8B
|
phosphodiesterase 8B
|
|
chr5_+_77210667
Show fit
|
2.70 |
ENST00000264917.10
|
PDE8B
|
phosphodiesterase 8B
|
|
chr8_+_1763766
Show fit
|
2.46 |
ENST00000635751.1
ENST00000331222.6
ENST00000637156.1
ENST00000636934.1
ENST00000637083.1
|
CLN8
|
CLN8 transmembrane ER and ERGIC protein
|
|
chr2_-_98936155
Show fit
|
2.29 |
ENST00000428096.5
ENST00000397899.7
ENST00000420294.1
|
CRACDL
|
CRACD like
|
|
chr13_+_34942263
Show fit
|
2.15 |
ENST00000379939.7
ENST00000400445.7
|
NBEA
|
neurobeachin
|
|
chr7_-_117873420
Show fit
|
2.14 |
ENST00000160373.8
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr8_+_1763832
Show fit
|
1.87 |
ENST00000520991.3
|
CLN8
|
CLN8 transmembrane ER and ERGIC protein
|
|
chr4_-_148442342
Show fit
|
1.85 |
ENST00000358102.8
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2
|
|
chr4_-_148442508
Show fit
|
1.82 |
ENST00000625323.2
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2
|
|
chr11_+_2444986
Show fit
|
1.76 |
ENST00000155840.12
|
KCNQ1
|
potassium voltage-gated channel subfamily Q member 1
|
|
chr8_+_1763752
Show fit
|
1.76 |
ENST00000519254.2
|
CLN8
|
CLN8 transmembrane ER and ERGIC protein
|
|
chr17_-_76141240
Show fit
|
1.68 |
ENST00000322957.7
|
FOXJ1
|
forkhead box J1
|
|
chr6_-_89412219
Show fit
|
1.59 |
ENST00000369415.9
|
RRAGD
|
Ras related GTP binding D
|
|
chr11_+_125951783
Show fit
|
1.58 |
ENST00000640497.1
|
VSIG10L2
|
V-set and immunoglobulin domain containing 10 like 2
|
|
chr6_-_89412069
Show fit
|
1.46 |
ENST00000359203.3
|
RRAGD
|
Ras related GTP binding D
|
|
chr4_-_148444674
Show fit
|
1.27 |
ENST00000344721.8
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2
|
|
chr11_-_67505323
Show fit
|
1.23 |
ENST00000356404.8
ENST00000436757.6
|
PITPNM1
|
phosphatidylinositol transfer protein membrane associated 1
|
|
chr6_-_29628038
Show fit
|
1.11 |
ENST00000355973.7
ENST00000377012.8
|
GABBR1
|
gamma-aminobutyric acid type B receptor subunit 1
|
|
chr13_+_24680407
Show fit
|
1.11 |
ENST00000381946.5
ENST00000218548.10
|
ATP12A
|
ATPase H+/K+ transporting non-gastric alpha2 subunit
|
|
chr19_+_13024917
Show fit
|
1.01 |
ENST00000587260.1
|
NFIX
|
nuclear factor I X
|
|
chr14_+_100381974
Show fit
|
0.97 |
ENST00000542471.2
|
WDR25
|
WD repeat domain 25
|
|
chr12_+_108129276
Show fit
|
0.88 |
ENST00000547525.6
|
WSCD2
|
WSC domain containing 2
|
|
chr19_-_14206168
Show fit
|
0.88 |
ENST00000361434.7
ENST00000340736.10
|
ADGRL1
|
adhesion G protein-coupled receptor L1
|
|
chr1_+_85062304
Show fit
|
0.83 |
ENST00000326813.12
ENST00000528899.5
ENST00000294664.11
|
DNAI3
|
dynein axonemal intermediate chain 3
|
|
chr11_-_2161158
Show fit
|
0.83 |
ENST00000421783.1
ENST00000397262.5
ENST00000381330.5
ENST00000250971.7
ENST00000397270.1
|
INS
INS-IGF2
|
insulin
INS-IGF2 readthrough
|
|
chr2_+_218323148
Show fit
|
0.82 |
ENST00000258362.7
|
PNKD
|
PNKD metallo-beta-lactamase domain containing
|
|
chr6_-_33069242
Show fit
|
0.81 |
ENST00000437811.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1
|
|
chr4_+_52051285
Show fit
|
0.79 |
ENST00000295213.9
ENST00000419395.6
|
SPATA18
|
spermatogenesis associated 18
|
|
chr17_-_80220325
Show fit
|
0.78 |
ENST00000326317.11
ENST00000570427.1
ENST00000570923.1
|
SGSH
|
N-sulfoglucosamine sulfohydrolase
|
|
chr2_+_10721649
Show fit
|
0.78 |
ENST00000381661.3
|
ATP6V1C2
|
ATPase H+ transporting V1 subunit C2
|
|
chr3_-_12158901
Show fit
|
0.76 |
ENST00000287814.5
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr11_+_1223053
Show fit
|
0.75 |
ENST00000529681.5
|
MUC5B
|
mucin 5B, oligomeric mucus/gel-forming
|
|
chr6_-_39229465
Show fit
|
0.72 |
ENST00000359534.4
|
KCNK5
|
potassium two pore domain channel subfamily K member 5
|
|
chr2_+_10721623
Show fit
|
0.71 |
ENST00000272238.9
|
ATP6V1C2
|
ATPase H+ transporting V1 subunit C2
|
|
chr5_+_76403266
Show fit
|
0.70 |
ENST00000274364.11
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr1_+_3690654
Show fit
|
0.68 |
ENST00000378285.5
ENST00000378280.5
ENST00000378288.8
|
TP73
|
tumor protein p73
|
|
chr17_-_5123102
Show fit
|
0.66 |
ENST00000250076.7
|
ZNF232
|
zinc finger protein 232
|
|
chr4_+_1011595
Show fit
|
0.65 |
ENST00000510644.6
|
FGFRL1
|
fibroblast growth factor receptor like 1
|
|
chr9_-_122227525
Show fit
|
0.65 |
ENST00000373755.6
ENST00000373754.6
|
LHX6
|
LIM homeobox 6
|
|
chr4_-_140427635
Show fit
|
0.64 |
ENST00000325617.10
ENST00000414773.5
|
CLGN
|
calmegin
|
|
chr17_+_80220406
Show fit
|
0.62 |
ENST00000573809.5
ENST00000361193.8
ENST00000574967.5
ENST00000576126.5
ENST00000411502.7
ENST00000546047.6
ENST00000572725.5
|
SLC26A11
|
solute carrier family 26 member 11
|
|
chr6_+_33200860
Show fit
|
0.61 |
ENST00000374677.8
|
SLC39A7
|
solute carrier family 39 member 7
|
|
chr17_+_40062810
Show fit
|
0.60 |
ENST00000584985.5
ENST00000264637.8
|
THRA
|
thyroid hormone receptor alpha
|
|
chr12_+_49539022
Show fit
|
0.59 |
ENST00000257981.7
|
KCNH3
|
potassium voltage-gated channel subfamily H member 3
|
|
chr8_+_67064316
Show fit
|
0.59 |
ENST00000675306.2
ENST00000678017.1
ENST00000262210.11
ENST00000675869.1
ENST00000677009.1
ENST00000676847.1
ENST00000676471.1
ENST00000678542.1
ENST00000677619.1
ENST00000676605.1
ENST00000678553.1
ENST00000674993.1
ENST00000678318.1
ENST00000676573.1
ENST00000676317.1
ENST00000677592.1
ENST00000679226.1
ENST00000675955.1
ENST00000676882.1
ENST00000678616.1
ENST00000678645.1
ENST00000678747.1
|
CSPP1
|
centrosome and spindle pole associated protein 1
|
|
chr10_+_49610297
Show fit
|
0.59 |
ENST00000374115.5
|
SLC18A3
|
solute carrier family 18 member A3
|
|
chr20_+_36573458
Show fit
|
0.57 |
ENST00000373874.6
|
TGIF2
|
TGFB induced factor homeobox 2
|
|
chr6_+_33200820
Show fit
|
0.56 |
ENST00000374675.7
|
SLC39A7
|
solute carrier family 39 member 7
|
|
chr6_+_143536811
Show fit
|
0.56 |
ENST00000367584.8
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chrX_-_54183207
Show fit
|
0.56 |
ENST00000375180.7
ENST00000328235.4
ENST00000477084.1
|
FAM120C
|
family with sequence similarity 120C
|
|
chrX_-_34657274
Show fit
|
0.55 |
ENST00000275954.4
|
TMEM47
|
transmembrane protein 47
|
|
chr16_+_67562453
Show fit
|
0.55 |
ENST00000646076.1
|
CTCF
|
CCCTC-binding factor
|
|
chr1_+_3652565
Show fit
|
0.54 |
ENST00000354437.8
ENST00000357733.7
ENST00000346387.8
|
TP73
|
tumor protein p73
|
|
chr19_-_3062464
Show fit
|
0.54 |
ENST00000327141.9
|
TLE5
|
TLE family member 5, transcriptional modulator
|
|
chr5_+_138352674
Show fit
|
0.53 |
ENST00000314358.10
|
KDM3B
|
lysine demethylase 3B
|
|
chr4_-_167234579
Show fit
|
0.53 |
ENST00000502330.5
ENST00000357154.7
ENST00000421836.6
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3
|
|
chr16_+_67562514
Show fit
|
0.53 |
ENST00000264010.10
ENST00000401394.6
ENST00000646771.1
|
CTCF
|
CCCTC-binding factor
|
|
chr4_-_167234426
Show fit
|
0.53 |
ENST00000541354.5
ENST00000509854.5
ENST00000512681.5
ENST00000357545.9
ENST00000510741.5
ENST00000510403.5
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3
|
|
chrX_+_24693879
Show fit
|
0.53 |
ENST00000379068.8
ENST00000677890.1
ENST00000379059.7
|
POLA1
|
DNA polymerase alpha 1, catalytic subunit
|
|
chr8_-_101205561
Show fit
|
0.52 |
ENST00000519744.5
ENST00000311212.9
ENST00000521272.5
ENST00000519882.5
|
ZNF706
|
zinc finger protein 706
|
|
chr9_-_115118198
Show fit
|
0.51 |
ENST00000534839.1
ENST00000535648.5
|
TNC
|
tenascin C
|
|
chr16_+_30985181
Show fit
|
0.51 |
ENST00000262520.10
ENST00000297679.10
ENST00000562932.5
ENST00000574447.1
|
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7
|
|
chr1_+_3652508
Show fit
|
0.50 |
ENST00000378295.9
ENST00000604074.5
|
TP73
|
tumor protein p73
|
|
chr7_+_23680130
Show fit
|
0.50 |
ENST00000409192.7
ENST00000409653.5
ENST00000409994.3
ENST00000344962.9
|
FAM221A
|
family with sequence similarity 221 member A
|
|
chr16_+_67807082
Show fit
|
0.49 |
ENST00000567852.5
ENST00000565148.5
ENST00000388833.7
ENST00000561654.5
ENST00000431934.2
|
TSNAXIP1
|
translin associated factor X interacting protein 1
|
|
chr16_+_67806765
Show fit
|
0.49 |
ENST00000415766.7
ENST00000561639.6
|
TSNAXIP1
|
translin associated factor X interacting protein 1
|
|
chr19_+_13795434
Show fit
|
0.48 |
ENST00000254323.6
|
ZSWIM4
|
zinc finger SWIM-type containing 4
|
|
chr5_+_134371561
Show fit
|
0.48 |
ENST00000265339.7
ENST00000506787.5
ENST00000507277.1
|
UBE2B
|
ubiquitin conjugating enzyme E2 B
|
|
chr12_-_51026325
Show fit
|
0.48 |
ENST00000547198.5
ENST00000643884.1
|
SLC11A2
|
solute carrier family 11 member 2
|
|
chr17_-_15999689
Show fit
|
0.47 |
ENST00000399277.6
|
ZSWIM7
|
zinc finger SWIM-type containing 7
|
|
chr1_-_154956086
Show fit
|
0.47 |
ENST00000368463.8
ENST00000368460.7
ENST00000368465.5
|
PBXIP1
|
PBX homeobox interacting protein 1
|
|
chr17_-_4560564
Show fit
|
0.47 |
ENST00000574584.1
ENST00000381550.8
ENST00000301395.7
|
GGT6
|
gamma-glutamyltransferase 6
|
|
chr21_+_32412648
Show fit
|
0.46 |
ENST00000401402.7
ENST00000382699.7
ENST00000300255.7
|
EVA1C
|
eva-1 homolog C
|
|
chr10_+_44959780
Show fit
|
0.43 |
ENST00000340258.10
ENST00000427758.5
|
RASSF4
|
Ras association domain family member 4
|
|
chr14_+_93185304
Show fit
|
0.42 |
ENST00000415050.3
|
TMEM251
|
transmembrane protein 251
|
|
chr5_-_180072086
Show fit
|
0.42 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130
|
|
chr17_-_65560296
Show fit
|
0.42 |
ENST00000585045.1
ENST00000611991.1
|
AXIN2
|
axin 2
|
|
chr12_-_48350771
Show fit
|
0.40 |
ENST00000544117.6
ENST00000548932.5
ENST00000549125.5
ENST00000301042.7
ENST00000547026.6
|
ZNF641
|
zinc finger protein 641
|
|
chr4_+_105895458
Show fit
|
0.40 |
ENST00000379987.7
|
NPNT
|
nephronectin
|
|
chr14_+_91114431
Show fit
|
0.39 |
ENST00000428926.6
ENST00000517362.5
|
DGLUCY
|
D-glutamate cyclase
|
|
chr14_+_91114364
Show fit
|
0.39 |
ENST00000518868.5
|
DGLUCY
|
D-glutamate cyclase
|
|
chr4_-_167234266
Show fit
|
0.39 |
ENST00000511269.5
ENST00000506697.5
ENST00000512042.1
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3
|
|
chr17_+_78232006
Show fit
|
0.39 |
ENST00000550981.7
ENST00000591033.2
|
TMEM235
|
transmembrane protein 235
|
|
chr9_-_137188540
Show fit
|
0.39 |
ENST00000323927.3
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr19_-_12881460
Show fit
|
0.39 |
ENST00000592506.1
|
DNASE2
|
deoxyribonuclease 2, lysosomal
|
|
chr17_-_5500997
Show fit
|
0.38 |
ENST00000568641.2
|
ENSG00000286190.2
|
novel protein
|
|
chr20_+_36573589
Show fit
|
0.38 |
ENST00000373872.9
ENST00000650844.1
|
TGIF2
|
TGFB induced factor homeobox 2
|
|
chr4_+_73853290
Show fit
|
0.38 |
ENST00000226524.4
|
PF4V1
|
platelet factor 4 variant 1
|
|
chr12_-_51026345
Show fit
|
0.38 |
ENST00000547732.5
ENST00000644495.1
ENST00000262052.9
ENST00000546488.5
ENST00000550714.5
ENST00000548193.5
ENST00000547579.5
ENST00000546743.5
|
SLC11A2
|
solute carrier family 11 member 2
|
|
chr21_+_41168142
Show fit
|
0.37 |
ENST00000330333.11
|
BACE2
|
beta-secretase 2
|
|
chr11_-_118152775
Show fit
|
0.37 |
ENST00000324727.9
|
SCN4B
|
sodium voltage-gated channel beta subunit 4
|
|
chr4_+_55346349
Show fit
|
0.37 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3
|
|
chr15_-_68229658
Show fit
|
0.37 |
ENST00000565471.6
ENST00000637494.1
ENST00000636314.1
ENST00000637667.1
ENST00000564752.1
ENST00000566347.5
ENST00000249806.11
ENST00000562767.2
|
CLN6
ENSG00000260007.3
|
CLN6 transmembrane ER protein
novel protein
|
|
chr16_-_18376839
Show fit
|
0.36 |
ENST00000427999.6
|
NPIPA9
|
nuclear pore complex interacting protein family, member A9
|
|
chr2_+_229922482
Show fit
|
0.36 |
ENST00000283946.8
ENST00000409992.1
|
FBXO36
|
F-box protein 36
|
|
chr17_-_10729973
Show fit
|
0.35 |
ENST00000578345.1
ENST00000341871.8
ENST00000455996.6
|
TMEM220
|
transmembrane protein 220
|
|
chr7_+_149873956
Show fit
|
0.35 |
ENST00000425642.3
ENST00000479613.5
ENST00000606024.5
ENST00000464662.5
|
ATP6V0E2
|
ATPase H+ transporting V0 subunit e2
|
|
chr12_-_58920465
Show fit
|
0.35 |
ENST00000320743.8
|
LRIG3
|
leucine rich repeats and immunoglobulin like domains 3
|
|
chr17_-_15999634
Show fit
|
0.35 |
ENST00000472495.5
|
ZSWIM7
|
zinc finger SWIM-type containing 7
|
|
chr17_+_20009320
Show fit
|
0.34 |
ENST00000679049.1
ENST00000472876.5
ENST00000583528.6
ENST00000680019.1
ENST00000681593.1
ENST00000395527.8
ENST00000583482.7
ENST00000679255.1
ENST00000681202.1
ENST00000677914.1
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr18_+_13218195
Show fit
|
0.33 |
ENST00000679167.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4
|
|
chr18_+_11752041
Show fit
|
0.32 |
ENST00000423027.8
|
GNAL
|
G protein subunit alpha L
|
|
chr22_-_46738205
Show fit
|
0.31 |
ENST00000216264.13
|
CERK
|
ceramide kinase
|
|
chr4_+_105895487
Show fit
|
0.31 |
ENST00000506666.5
ENST00000503451.5
|
NPNT
|
nephronectin
|
|
chr12_-_51026141
Show fit
|
0.31 |
ENST00000541174.6
|
SLC11A2
|
solute carrier family 11 member 2
|
|
chr11_-_119340816
Show fit
|
0.31 |
ENST00000528368.3
|
C1QTNF5
|
C1q and TNF related 5
|
|
chr11_+_73308237
Show fit
|
0.31 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17
|
|
chr6_-_39115145
Show fit
|
0.31 |
ENST00000229903.5
|
SAYSD1
|
SAYSVFN motif domain containing 1
|
|
chr12_+_51391273
Show fit
|
0.31 |
ENST00000535225.6
ENST00000358657.7
|
SLC4A8
|
solute carrier family 4 member 8
|
|
chr1_-_32336224
Show fit
|
0.31 |
ENST00000329421.8
|
MARCKSL1
|
MARCKS like 1
|
|
chr16_+_3012915
Show fit
|
0.30 |
ENST00000445369.3
|
CLDN9
|
claudin 9
|
|
chr1_+_113929304
Show fit
|
0.30 |
ENST00000426820.7
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr2_+_237859615
Show fit
|
0.30 |
ENST00000409726.5
ENST00000254661.5
|
RAMP1
|
receptor activity modifying protein 1
|
|
chr19_-_291132
Show fit
|
0.29 |
ENST00000327790.7
|
PLPP2
|
phospholipid phosphatase 2
|
|
chr1_-_5992407
Show fit
|
0.29 |
ENST00000378156.9
ENST00000622020.4
|
NPHP4
|
nephrocystin 4
|
|
chr5_-_9546066
Show fit
|
0.29 |
ENST00000382496.10
ENST00000652226.1
|
SEMA5A
|
semaphorin 5A
|
|
chr11_-_65382632
Show fit
|
0.29 |
ENST00000294187.10
ENST00000398802.6
ENST00000530936.1
|
SLC25A45
|
solute carrier family 25 member 45
|
|
chr17_-_15999589
Show fit
|
0.28 |
ENST00000486655.5
|
ZSWIM7
|
zinc finger SWIM-type containing 7
|
|
chr22_+_29306582
Show fit
|
0.28 |
ENST00000616432.4
ENST00000416823.1
ENST00000428622.1
|
GAS2L1
|
growth arrest specific 2 like 1
|
|
chr4_+_499194
Show fit
|
0.28 |
ENST00000453061.7
ENST00000310340.9
ENST00000504346.5
ENST00000503111.5
ENST00000383028.8
ENST00000509768.1
|
PIGG
|
phosphatidylinositol glycan anchor biosynthesis class G
|
|
chr1_+_89524871
Show fit
|
0.28 |
ENST00000639264.1
|
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B
|
|
chr6_-_39314445
Show fit
|
0.28 |
ENST00000453413.2
|
KCNK17
|
potassium two pore domain channel subfamily K member 17
|
|
chr11_-_62709493
Show fit
|
0.28 |
ENST00000405837.5
ENST00000531524.5
ENST00000524862.6
ENST00000679883.1
|
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin
|
|
chr18_+_11751494
Show fit
|
0.28 |
ENST00000269162.9
|
GNAL
|
G protein subunit alpha L
|
|
chr7_-_149773548
Show fit
|
0.28 |
ENST00000302017.4
|
ZNF467
|
zinc finger protein 467
|
|
chr12_-_7091873
Show fit
|
0.28 |
ENST00000538050.5
ENST00000536053.6
|
C1R
|
complement C1r
|
|
chr19_-_291365
Show fit
|
0.28 |
ENST00000591572.2
ENST00000269812.7
ENST00000633125.1
ENST00000434325.7
|
PLPP2
|
phospholipid phosphatase 2
|
|
chr10_+_132332273
Show fit
|
0.28 |
ENST00000368613.8
|
LRRC27
|
leucine rich repeat containing 27
|
|
chr1_+_89524819
Show fit
|
0.27 |
ENST00000439853.6
ENST00000330947.7
ENST00000449440.5
ENST00000640258.1
|
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B
|
|
chr19_+_48619291
Show fit
|
0.27 |
ENST00000340932.7
ENST00000601712.5
ENST00000600537.5
|
SPHK2
|
sphingosine kinase 2
|
|
chr4_-_499102
Show fit
|
0.26 |
ENST00000338977.5
ENST00000511833.3
|
ZNF721
|
zinc finger protein 721
|
|
chr11_-_126062782
Show fit
|
0.26 |
ENST00000531738.6
|
CDON
|
cell adhesion associated, oncogene regulated
|
|
chr19_+_48619528
Show fit
|
0.26 |
ENST00000598088.5
|
SPHK2
|
sphingosine kinase 2
|
|
chr21_-_33479914
Show fit
|
0.26 |
ENST00000542230.7
|
TMEM50B
|
transmembrane protein 50B
|
|
chr13_-_94712505
Show fit
|
0.26 |
ENST00000376945.4
|
SOX21
|
SRY-box transcription factor 21
|
|
chr17_-_17206264
Show fit
|
0.26 |
ENST00000321560.4
|
PLD6
|
phospholipase D family member 6
|
|
chr4_-_40629842
Show fit
|
0.26 |
ENST00000295971.12
|
RBM47
|
RNA binding motif protein 47
|
|
chr19_+_48619489
Show fit
|
0.25 |
ENST00000245222.9
|
SPHK2
|
sphingosine kinase 2
|
|
chr3_-_17742498
Show fit
|
0.25 |
ENST00000429383.8
ENST00000446863.5
ENST00000414349.5
ENST00000428355.5
ENST00000425944.5
ENST00000445294.5
ENST00000444471.5
ENST00000415814.6
|
TBC1D5
|
TBC1 domain family member 5
|
|
chr1_+_26111139
Show fit
|
0.25 |
ENST00000619836.4
ENST00000444713.5
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr19_+_1266653
Show fit
|
0.25 |
ENST00000586472.5
ENST00000589266.5
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr5_+_176810498
Show fit
|
0.25 |
ENST00000509580.2
|
UNC5A
|
unc-5 netrin receptor A
|
|
chr17_+_79034185
Show fit
|
0.25 |
ENST00000581774.5
|
C1QTNF1
|
C1q and TNF related 1
|
|
chrX_+_107628428
Show fit
|
0.25 |
ENST00000643795.2
ENST00000372418.4
ENST00000646815.1
ENST00000372435.10
ENST00000372419.3
ENST00000676092.1
|
PRPS1
|
phosphoribosyl pyrophosphate synthetase 1
|
|
chr4_+_3074661
Show fit
|
0.25 |
ENST00000355072.11
|
HTT
|
huntingtin
|
|
chr17_-_41034871
Show fit
|
0.25 |
ENST00000344363.7
|
KRTAP1-3
|
keratin associated protein 1-3
|
|
chr2_+_24491239
Show fit
|
0.25 |
ENST00000348332.8
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr5_+_176810552
Show fit
|
0.25 |
ENST00000329542.9
|
UNC5A
|
unc-5 netrin receptor A
|
|
chr1_-_54406385
Show fit
|
0.25 |
ENST00000610401.5
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr14_+_56579782
Show fit
|
0.24 |
ENST00000261556.11
ENST00000538838.5
|
TMEM260
|
transmembrane protein 260
|
|
chr15_+_65530754
Show fit
|
0.24 |
ENST00000566074.5
|
HACD3
|
3-hydroxyacyl-CoA dehydratase 3
|
|
chr19_+_33194308
Show fit
|
0.24 |
ENST00000253193.9
|
LRP3
|
LDL receptor related protein 3
|
|
chr9_-_83063159
Show fit
|
0.24 |
ENST00000340717.4
|
RASEF
|
RAS and EF-hand domain containing
|
|
chr3_-_64268161
Show fit
|
0.24 |
ENST00000564377.6
|
PRICKLE2
|
prickle planar cell polarity protein 2
|
|
chr11_-_76381053
Show fit
|
0.24 |
ENST00000260045.8
|
THAP12
|
THAP domain containing 12
|
|
chr2_-_45009401
Show fit
|
0.24 |
ENST00000303077.7
|
SIX2
|
SIX homeobox 2
|
|
chr13_+_38687068
Show fit
|
0.23 |
ENST00000280481.9
|
FREM2
|
FRAS1 related extracellular matrix 2
|
|
chr1_+_52056284
Show fit
|
0.23 |
ENST00000313334.13
ENST00000472944.6
ENST00000484036.1
|
BTF3L4
|
basic transcription factor 3 like 4
|
|
chr19_-_12881429
Show fit
|
0.23 |
ENST00000222219.8
|
DNASE2
|
deoxyribonuclease 2, lysosomal
|
|
chr16_-_8868687
Show fit
|
0.23 |
ENST00000618335.4
ENST00000570125.5
|
CARHSP1
|
calcium regulated heat stable protein 1
|
|
chr6_-_41705813
Show fit
|
0.23 |
ENST00000419574.6
ENST00000445214.2
|
TFEB
|
transcription factor EB
|
|
chr19_+_49114324
Show fit
|
0.23 |
ENST00000391864.7
|
LIN7B
|
lin-7 homolog B, crumbs cell polarity complex component
|
|
chr9_-_111484353
Show fit
|
0.23 |
ENST00000338205.9
ENST00000684092.1
|
ECPAS
|
Ecm29 proteasome adaptor and scaffold
|
|
chr14_+_91114388
Show fit
|
0.22 |
ENST00000519019.5
ENST00000523816.5
ENST00000517518.5
|
DGLUCY
|
D-glutamate cyclase
|
|
chr20_+_44885679
Show fit
|
0.22 |
ENST00000353703.9
ENST00000372839.7
ENST00000428262.1
ENST00000445830.1
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta
|
|
chr10_-_98268186
Show fit
|
0.22 |
ENST00000260702.4
|
LOXL4
|
lysyl oxidase like 4
|
|
chr7_-_100573865
Show fit
|
0.22 |
ENST00000622764.3
|
SAP25
|
Sin3A associated protein 25
|
|
chr19_+_49114352
Show fit
|
0.22 |
ENST00000221459.7
ENST00000486217.2
|
LIN7B
|
lin-7 homolog B, crumbs cell polarity complex component
|
|
chr12_+_55681647
Show fit
|
0.22 |
ENST00000614691.1
|
METTL7B
|
methyltransferase like 7B
|
|
chr15_+_65530418
Show fit
|
0.22 |
ENST00000562901.5
ENST00000261875.10
ENST00000442729.6
ENST00000565299.5
ENST00000568793.5
|
HACD3
|
3-hydroxyacyl-CoA dehydratase 3
|
|
chr5_-_111756245
Show fit
|
0.22 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein
|
|
chr1_+_26111798
Show fit
|
0.21 |
ENST00000374269.2
ENST00000374271.8
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr14_-_67533720
Show fit
|
0.21 |
ENST00000554278.6
|
TMEM229B
|
transmembrane protein 229B
|
|
chr16_-_8868994
Show fit
|
0.21 |
ENST00000311052.10
ENST00000565287.5
ENST00000611932.4
|
CARHSP1
|
calcium regulated heat stable protein 1
|
|
chr10_+_69180226
Show fit
|
0.21 |
ENST00000359655.9
ENST00000422378.1
|
SUPV3L1
|
Suv3 like RNA helicase
|
|
chr17_-_57988179
Show fit
|
0.21 |
ENST00000581208.2
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr12_+_55681711
Show fit
|
0.21 |
ENST00000394252.4
|
METTL7B
|
methyltransferase like 7B
|
|
chr4_+_55346213
Show fit
|
0.21 |
ENST00000679836.1
ENST00000264228.9
ENST00000679707.1
|
SRD5A3
ENSG00000288695.1
|
steroid 5 alpha-reductase 3
novel protein, SRD5A3-RP11-177J6.1 readthrough
|
|
chr11_-_67508091
Show fit
|
0.20 |
ENST00000531506.1
|
CDK2AP2
|
cyclin dependent kinase 2 associated protein 2
|
|
chr8_-_79767843
Show fit
|
0.20 |
ENST00000337919.9
|
HEY1
|
hes related family bHLH transcription factor with YRPW motif 1
|
|
chr9_+_128818491
Show fit
|
0.20 |
ENST00000372642.5
|
ENDOG
|
endonuclease G
|
|
chr11_+_59172116
Show fit
|
0.20 |
ENST00000227451.4
|
DTX4
|
deltex E3 ubiquitin ligase 4
|
|
chr7_+_150991005
Show fit
|
0.20 |
ENST00000297494.8
|
NOS3
|
nitric oxide synthase 3
|
|
chr3_+_23805941
Show fit
|
0.20 |
ENST00000306627.8
ENST00000346855.7
|
UBE2E1
|
ubiquitin conjugating enzyme E2 E1
|
|
chr19_-_9913819
Show fit
|
0.20 |
ENST00000593091.2
|
OLFM2
|
olfactomedin 2
|
|
chr10_+_38010617
Show fit
|
0.20 |
ENST00000307441.13
ENST00000374618.7
ENST00000628825.2
ENST00000458705.6
ENST00000432900.7
ENST00000469037.2
|
ZNF33A
|
zinc finger protein 33A
|
|
chr13_-_51804094
Show fit
|
0.20 |
ENST00000280056.6
|
DHRS12
|
dehydrogenase/reductase 12
|
|
chr18_+_13217676
Show fit
|
0.20 |
ENST00000679177.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4
|
|
chr21_+_45405117
Show fit
|
0.20 |
ENST00000651438.1
|
COL18A1
|
collagen type XVIII alpha 1 chain
|
|
chr1_+_244835616
Show fit
|
0.20 |
ENST00000366528.3
ENST00000411948.7
|
COX20
|
cytochrome c oxidase assembly factor COX20
|
|
chr17_-_39688016
Show fit
|
0.20 |
ENST00000579146.5
ENST00000300658.9
ENST00000378011.8
ENST00000429199.6
|
PGAP3
|
post-GPI attachment to proteins phospholipase 3
|
|
chr19_+_50358571
Show fit
|
0.20 |
ENST00000652203.1
|
NR1H2
|
nuclear receptor subfamily 1 group H member 2
|
|
chr16_-_8868343
Show fit
|
0.20 |
ENST00000562843.5
ENST00000561530.5
ENST00000396593.6
|
CARHSP1
|
calcium regulated heat stable protein 1
|
|
chr8_-_27605271
Show fit
|
0.20 |
ENST00000522098.1
|
CLU
|
clusterin
|
|
chr11_-_74398378
Show fit
|
0.19 |
ENST00000298198.5
|
PGM2L1
|
phosphoglucomutase 2 like 1
|
|
chrX_+_153517672
Show fit
|
0.19 |
ENST00000349466.6
ENST00000370186.5
ENST00000359149.8
|
ATP2B3
|
ATPase plasma membrane Ca2+ transporting 3
|
|
chr9_-_83063135
Show fit
|
0.19 |
ENST00000376447.4
|
RASEF
|
RAS and EF-hand domain containing
|