Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
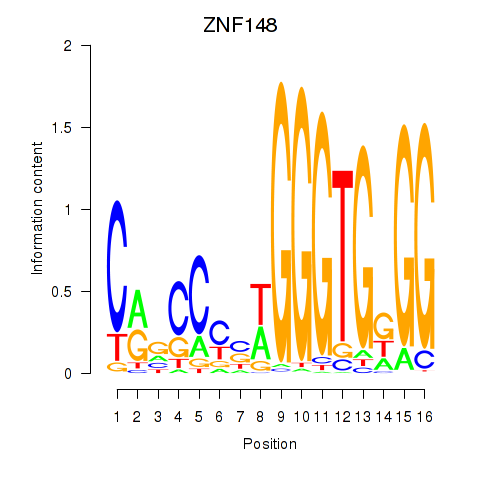
Results for ZNF148
Z-value: 1.49
Transcription factors associated with ZNF148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF148
|
ENSG00000163848.20 | ZNF148 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF148 | hg38_v1_chr3_-_125375249_125375330 | -0.41 | 2.3e-02 | Click! |
Activity profile of ZNF148 motif
Sorted Z-values of ZNF148 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF148
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 1.5 | 4.6 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 1.3 | 3.8 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 1.0 | 3.9 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.0 | 8.7 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 1.0 | 6.7 | GO:0060068 | vagina development(GO:0060068) |
| 0.8 | 2.4 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.8 | 3.8 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.7 | 3.7 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.7 | 13.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.7 | 2.2 | GO:0070650 | endoplasmic reticulum polarization(GO:0061163) actin filament bundle retrograde transport(GO:0061573) actin filament bundle distribution(GO:0070650) |
| 0.6 | 4.5 | GO:0030421 | defecation(GO:0030421) |
| 0.6 | 2.4 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.6 | 4.1 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.6 | 13.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.5 | 2.7 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.5 | 3.7 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.5 | 3.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.5 | 1.5 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.5 | 1.9 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.5 | 7.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.4 | 1.8 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.4 | 7.1 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.4 | 0.9 | GO:0035789 | cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.4 | 2.6 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.4 | 3.4 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.4 | 1.2 | GO:1904604 | regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 0.4 | 3.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.4 | 3.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.4 | 2.4 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.3 | 2.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.3 | 1.7 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.3 | 1.7 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.3 | 1.0 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.3 | 2.2 | GO:0072103 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.3 | 0.9 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.3 | 1.2 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.3 | 1.2 | GO:0030047 | actin modification(GO:0030047) |
| 0.3 | 3.0 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 2.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.2 | 0.7 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 | 3.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 3.1 | GO:0060083 | response to carbon monoxide(GO:0034465) smooth muscle contraction involved in micturition(GO:0060083) |
| 0.2 | 1.4 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.2 | 6.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.7 | GO:1902269 | putrescine transport(GO:0015847) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 6.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 2.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 0.9 | GO:0048627 | myoblast development(GO:0048627) |
| 0.2 | 2.4 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.2 | 1.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 12.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 4.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.2 | 0.2 | GO:2000137 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.2 | 2.1 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.2 | 0.6 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.2 | 0.7 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.2 | 0.5 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 2.7 | GO:1903764 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.2 | 0.8 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 1.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 2.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 0.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.4 | GO:1901874 | negative regulation of post-translational protein modification(GO:1901874) |
| 0.1 | 1.5 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 0.9 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 2.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.4 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 2.1 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 1.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 1.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 2.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.7 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.1 | 1.4 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 1.2 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.9 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.8 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.1 | 1.9 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 1.2 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.1 | 1.5 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.9 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) |
| 0.1 | 1.2 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 0.7 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.3 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.4 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.1 | 0.3 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.1 | 0.5 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.9 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 1.8 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:0035048 | splicing factor protein import into nucleus(GO:0035048) |
| 0.1 | 1.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 | 1.7 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 1.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.4 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.5 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.1 | 3.6 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 0.9 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 2.2 | GO:0014041 | regulation of neuron maturation(GO:0014041) |
| 0.1 | 1.5 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 0.6 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 | 0.3 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.6 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.1 | 5.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.7 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 2.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 3.0 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.5 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 0.5 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 1.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 1.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.9 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 1.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.1 | 0.8 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 1.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.3 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 4.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 2.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 2.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 1.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 3.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.6 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.5 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 1.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 1.6 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 2.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.2 | GO:0002879 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 0.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.3 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 1.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) diapedesis(GO:0050904) |
| 0.1 | 0.3 | GO:0072366 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 | 0.2 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.1 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 1.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 1.0 | GO:0021696 | cerebellar cortex morphogenesis(GO:0021696) |
| 0.0 | 3.7 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 1.2 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 0.9 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.2 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0014848 | urinary bladder smooth muscle contraction(GO:0014832) urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 4.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.6 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.8 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.3 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.8 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.1 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.0 | 1.6 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.4 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.4 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.9 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 | 0.7 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.2 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.9 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 1.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.7 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.4 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 2.7 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.3 | GO:0097647 | positive regulation of protein kinase A signaling(GO:0010739) dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.5 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.3 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.0 | 0.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.4 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 1.1 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.5 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 1.2 | GO:0032642 | regulation of chemokine production(GO:0032642) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 1.1 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 1.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 1.4 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0014061 | regulation of norepinephrine secretion(GO:0014061) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 2.3 | GO:0017156 | calcium ion regulated exocytosis(GO:0017156) |
| 0.0 | 0.5 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.3 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.1 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.0 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.2 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.7 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.6 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.6 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.5 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.9 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.5 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.7 | GO:0032449 | CBM complex(GO:0032449) |
| 0.9 | 4.6 | GO:0031523 | Myb complex(GO:0031523) |
| 0.7 | 3.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.6 | 11.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.5 | 13.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 2.4 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.4 | 1.7 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 2.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 3.6 | GO:0043256 | laminin complex(GO:0043256) |
| 0.3 | 5.6 | GO:0033643 | host cell part(GO:0033643) |
| 0.2 | 2.5 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.2 | 1.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.2 | 4.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 3.8 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.2 | 3.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.2 | 0.6 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 0.5 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 0.9 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 2.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.5 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.1 | 0.6 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 0.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 9.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 2.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.8 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.7 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 2.9 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 1.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.7 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 8.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.7 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 8.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.2 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.1 | 1.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 4.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 4.0 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 1.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.3 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 1.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.0 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.0 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 4.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 1.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 3.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.8 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 1.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 3.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 2.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 1.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 2.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 9.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 1.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.1 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 2.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.6 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.9 | 3.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.9 | 2.6 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.6 | 7.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.6 | 6.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.6 | 1.8 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.5 | 3.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.5 | 3.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.4 | 1.2 | GO:1904599 | advanced glycation end-product binding(GO:1904599) |
| 0.4 | 2.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.4 | 3.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 7.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 5.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 1.2 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 3.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.3 | 15.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.3 | 0.9 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.3 | 0.9 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.3 | 1.5 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.3 | 1.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 1.5 | GO:0042835 | BRE binding(GO:0042835) |
| 0.2 | 2.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 4.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 3.9 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.2 | 2.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.2 | 2.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 0.7 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.2 | 2.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 2.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 1.0 | GO:0030395 | lactose binding(GO:0030395) |
| 0.2 | 0.6 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.2 | 0.6 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.2 | 1.8 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.2 | 4.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 4.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 3.9 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 1.8 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) PH domain binding(GO:0042731) |
| 0.2 | 2.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 1.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 1.6 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 12.0 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 9.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 2.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.5 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 1.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.4 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.1 | 3.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.3 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 1.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 3.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.8 | GO:0016413 | carnitine O-acyltransferase activity(GO:0016406) O-acetyltransferase activity(GO:0016413) |
| 0.1 | 2.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.5 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 1.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.3 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 2.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 3.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 3.3 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.1 | 2.4 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.1 | 0.7 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 2.1 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 3.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 2.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 1.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.6 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 0.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 1.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.4 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 1.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 1.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.5 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 6.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 2.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 8.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.4 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 2.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 2.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 5.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.2 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.8 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 3.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 4.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.1 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 2.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 5.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 5.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 6.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.5 | GO:0017016 | Ras GTPase binding(GO:0017016) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 3.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 5.4 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 2.4 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 3.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 6.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 6.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 6.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 6.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 3.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 1.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 15.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 3.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 7.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 10.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 5.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 5.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 4.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 3.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 5.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 4.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 2.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 0.8 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 3.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 3.2 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 3.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 0.9 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 3.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 2.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 1.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.0 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 2.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.7 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 2.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 3.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 5.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 3.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 2.6 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.8 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |