Project
Mucociliary differentiation, bronchial epithelial cells, human (Ross 2007)
Navigation
Downloads
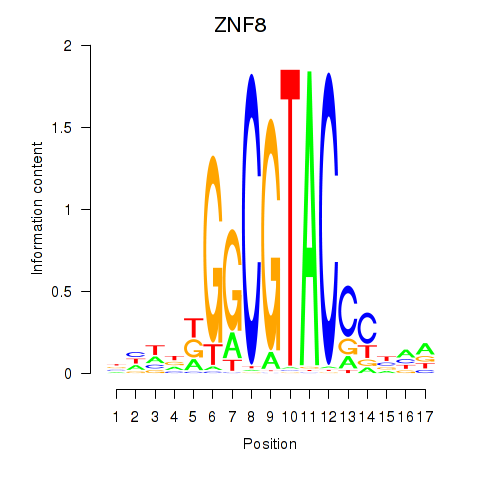
Results for ZNF8
Z-value: 0.44
Transcription factors associated with ZNF8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF8
|
ENSG00000278129.2 | ZNF8 |
|
ZNF8
|
ENSG00000278129.2 | ZNF8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF8 | hg38_v1_chr19_+_58278948_58278992 | 0.51 | 4.2e-03 | Click! |
Activity profile of ZNF8 motif
Sorted Z-values of ZNF8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_87407965 | 2.13 |
ENST00000369562.9
|
CFAP206
|
cilia and flagella associated protein 206 |
| chr4_+_185426234 | 1.97 |
ENST00000511138.5
ENST00000511581.5 ENST00000378850.5 |
C4orf47
|
chromosome 4 open reading frame 47 |
| chr7_+_48035511 | 1.96 |
ENST00000420324.5
ENST00000539619.5 ENST00000435376.5 ENST00000430738.5 ENST00000348904.4 |
C7orf57
|
chromosome 7 open reading frame 57 |
| chr2_+_119431846 | 1.80 |
ENST00000306406.5
|
TMEM37
|
transmembrane protein 37 |
| chr3_-_9952337 | 1.23 |
ENST00000411976.2
ENST00000412055.6 |
PRRT3
|
proline rich transmembrane protein 3 |
| chr3_+_113838772 | 1.12 |
ENST00000358160.9
|
GRAMD1C
|
GRAM domain containing 1C |
| chr6_+_29723340 | 1.03 |
ENST00000334668.8
|
HLA-F
|
major histocompatibility complex, class I, F |
| chr6_+_29723421 | 1.00 |
ENST00000259951.12
ENST00000434407.6 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr6_+_18155399 | 0.79 |
ENST00000650836.2
ENST00000449850.2 ENST00000297792.9 |
KDM1B
|
lysine demethylase 1B |
| chr9_+_1050330 | 0.60 |
ENST00000382255.7
ENST00000382251.7 ENST00000412350.6 ENST00000358146.7 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr13_+_36819214 | 0.51 |
ENST00000255476.3
|
RFXAP
|
regulatory factor X associated protein |
| chr6_-_26056460 | 0.50 |
ENST00000343677.4
|
H1-2
|
H1.2 linker histone, cluster member |
| chr16_+_29962049 | 0.34 |
ENST00000279396.11
ENST00000575829.6 ENST00000561899.6 |
TMEM219
|
transmembrane protein 219 |
| chr9_+_93263948 | 0.33 |
ENST00000448251.5
|
WNK2
|
WNK lysine deficient protein kinase 2 |
| chr20_+_38748448 | 0.33 |
ENST00000243903.6
|
ACTR5
|
actin related protein 5 |
| chr5_-_94111627 | 0.30 |
ENST00000505869.5
ENST00000395965.8 ENST00000509163.5 |
FAM172A
|
family with sequence similarity 172 member A |
| chr11_-_46617170 | 0.30 |
ENST00000326737.3
|
HARBI1
|
harbinger transposase derived 1 |
| chr1_-_186375671 | 0.28 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr4_+_169620527 | 0.27 |
ENST00000360642.7
ENST00000512813.5 ENST00000513761.6 |
CLCN3
|
chloride voltage-gated channel 3 |
| chr5_-_138575359 | 0.25 |
ENST00000297185.9
ENST00000678300.1 ENST00000677425.1 ENST00000677064.1 ENST00000507115.6 |
HSPA9
|
heat shock protein family A (Hsp70) member 9 |
| chr4_+_169620509 | 0.25 |
ENST00000347613.8
|
CLCN3
|
chloride voltage-gated channel 3 |
| chr2_+_171434196 | 0.24 |
ENST00000375255.8
ENST00000539783.5 |
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr5_+_103120264 | 0.24 |
ENST00000358359.8
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr22_-_38656353 | 0.23 |
ENST00000535113.7
|
FAM227A
|
family with sequence similarity 227 member A |
| chr13_+_52652828 | 0.20 |
ENST00000310528.9
ENST00000343788.10 |
SUGT1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr1_-_54801161 | 0.19 |
ENST00000371274.8
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr6_-_18155053 | 0.18 |
ENST00000309983.5
|
TPMT
|
thiopurine S-methyltransferase |
| chr2_-_239400991 | 0.17 |
ENST00000543185.6
|
HDAC4
|
histone deacetylase 4 |
| chr2_-_239400949 | 0.16 |
ENST00000345617.7
|
HDAC4
|
histone deacetylase 4 |
| chr2_+_32357009 | 0.15 |
ENST00000421745.6
|
BIRC6
|
baculoviral IAP repeat containing 6 |
| chr1_+_186375813 | 0.15 |
ENST00000419367.8
ENST00000287859.11 ENST00000367470.8 |
ODR4
|
odr-4 GPCR localization factor homolog |
| chr22_+_29767351 | 0.15 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chrX_+_129779930 | 0.12 |
ENST00000356892.4
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr9_-_37576365 | 0.11 |
ENST00000432825.7
|
FBXO10
|
F-box protein 10 |
| chr2_+_197500398 | 0.11 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr16_+_68539213 | 0.11 |
ENST00000570495.5
ENST00000564323.5 ENST00000562156.5 ENST00000573685.5 ENST00000563169.7 ENST00000611381.4 |
ZFP90
|
ZFP90 zinc finger protein |
| chr1_-_54801290 | 0.11 |
ENST00000371276.9
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr9_-_32552553 | 0.09 |
ENST00000379858.1
ENST00000360538.7 ENST00000681750.1 ENST00000680198.1 |
TOPORS
ENSG00000288684.1
|
TOP1 binding arginine/serine rich protein, E3 ubiquitin ligase novel protein |
| chr19_-_18281612 | 0.09 |
ENST00000252818.5
|
JUND
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr22_-_29766934 | 0.08 |
ENST00000344318.4
|
ZMAT5
|
zinc finger matrin-type 5 |
| chr2_+_197500371 | 0.08 |
ENST00000409468.1
ENST00000233893.10 |
HSPE1
|
heat shock protein family E (Hsp10) member 1 |
| chr5_+_139341875 | 0.08 |
ENST00000511706.5
|
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr19_+_37507129 | 0.08 |
ENST00000586138.5
ENST00000588578.5 ENST00000587986.5 |
ZNF793
|
zinc finger protein 793 |
| chr17_-_58514617 | 0.08 |
ENST00000682306.1
|
MTMR4
|
myotubularin related protein 4 |
| chr1_+_25338294 | 0.07 |
ENST00000374358.5
|
TMEM50A
|
transmembrane protein 50A |
| chr3_+_49199477 | 0.07 |
ENST00000452691.7
ENST00000366429.2 |
IHO1
|
interactor of HORMAD1 1 |
| chr17_+_77185210 | 0.07 |
ENST00000431431.6
|
SEC14L1
|
SEC14 like lipid binding 1 |
| chr5_+_139341826 | 0.07 |
ENST00000265192.9
|
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr12_-_122266425 | 0.07 |
ENST00000643696.1
ENST00000267199.9 |
VPS33A
|
VPS33A core subunit of CORVET and HOPS complexes |
| chr16_+_21953341 | 0.06 |
ENST00000268379.9
ENST00000561553.5 ENST00000565331.5 |
UQCRC2
|
ubiquinol-cytochrome c reductase core protein 2 |
| chr1_+_70205680 | 0.05 |
ENST00000370951.5
|
SRSF11
|
serine and arginine rich splicing factor 11 |
| chr16_-_53503192 | 0.04 |
ENST00000568596.5
ENST00000394657.12 ENST00000570004.5 ENST00000564497.1 ENST00000300245.8 |
AKTIP
|
AKT interacting protein |
| chr13_+_21176629 | 0.03 |
ENST00000309594.5
|
MRPL57
|
mitochondrial ribosomal protein L57 |
| chr17_+_50508384 | 0.03 |
ENST00000436259.6
ENST00000652471.1 ENST00000323776.11 ENST00000419930.2 |
MYCBPAP
|
MYCBP associated protein |
| chr6_+_44246166 | 0.02 |
ENST00000620073.4
|
HSP90AB1
|
heat shock protein 90 alpha family class B member 1 |
| chr1_+_70205708 | 0.02 |
ENST00000370950.7
|
SRSF11
|
serine and arginine rich splicing factor 11 |
| chr12_+_52069967 | 0.02 |
ENST00000336854.9
ENST00000550604.1 ENST00000553049.5 ENST00000548915.1 |
ATG101
|
autophagy related 101 |
| chr1_-_228109255 | 0.01 |
ENST00000483159.1
ENST00000366744.5 ENST00000348259.9 ENST00000495434.1 ENST00000295008.8 ENST00000366731.9 ENST00000366746.7 ENST00000366747.7 ENST00000336520.8 ENST00000391867.8 ENST00000457264.5 ENST00000464148.2 ENST00000336300.9 ENST00000430433.5 |
MRPL55
|
mitochondrial ribosomal protein L55 |
| chr17_+_35809474 | 0.01 |
ENST00000604879.5
ENST00000603427.5 ENST00000603777.5 ENST00000605844.6 ENST00000604841.5 |
TAF15
|
TATA-box binding protein associated factor 15 |
| chr2_-_105329685 | 0.01 |
ENST00000393359.7
|
TGFBRAP1
|
transforming growth factor beta receptor associated protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.6 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.3 | GO:0031445 | regulation of heterochromatin assembly(GO:0031445) positive regulation of heterochromatin assembly(GO:0031453) |
| 0.1 | 0.8 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 2.1 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.2 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.3 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0001534 | radial spoke(GO:0001534) axonemal outer doublet(GO:0097545) |
| 0.2 | 2.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.3 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.8 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |