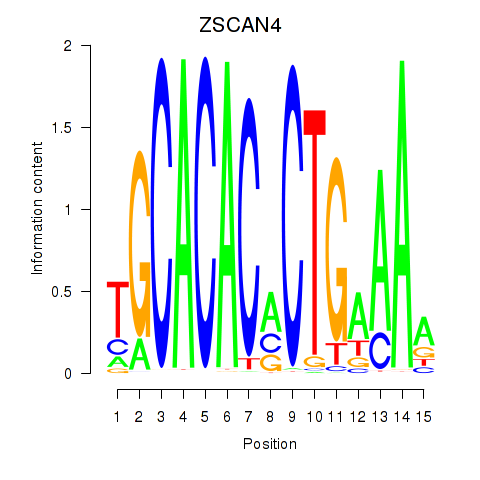
|
chr11_-_125592448
Show fit
|
1.15 |
ENST00000648911.1
|
FEZ1
|
fasciculation and elongation protein zeta 1
|
|
chr8_-_23854796
Show fit
|
1.14 |
ENST00000290271.7
|
STC1
|
stanniocalcin 1
|
|
chr9_-_122228845
Show fit
|
1.09 |
ENST00000394319.8
ENST00000340587.7
|
LHX6
|
LIM homeobox 6
|
|
chr8_-_121641424
Show fit
|
1.00 |
ENST00000303924.5
|
HAS2
|
hyaluronan synthase 2
|
|
chr19_-_17405554
Show fit
|
0.89 |
ENST00000252593.7
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr5_+_31193678
Show fit
|
0.78 |
ENST00000265071.3
|
CDH6
|
cadherin 6
|
|
chr4_-_103198331
Show fit
|
0.74 |
ENST00000265148.9
ENST00000514974.1
|
CENPE
|
centromere protein E
|
|
chr11_-_125496122
Show fit
|
0.72 |
ENST00000527534.1
ENST00000278919.8
ENST00000366139.3
|
FEZ1
|
fasciculation and elongation protein zeta 1
|
|
chrX_-_20116871
Show fit
|
0.70 |
ENST00000379651.7
ENST00000443379.7
ENST00000379643.10
|
MAP7D2
|
MAP7 domain containing 2
|
|
chr3_-_195811889
Show fit
|
0.67 |
ENST00000475231.5
|
MUC4
|
mucin 4, cell surface associated
|
|
chr12_+_40692413
Show fit
|
0.65 |
ENST00000551295.7
ENST00000547702.5
ENST00000551424.5
|
CNTN1
|
contactin 1
|
|
chr9_+_125748175
Show fit
|
0.64 |
ENST00000491787.7
ENST00000447726.6
|
PBX3
|
PBX homeobox 3
|
|
chr3_+_98763331
Show fit
|
0.62 |
ENST00000485391.5
ENST00000492254.5
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6
|
|
chr2_+_240625237
Show fit
|
0.62 |
ENST00000407714.1
|
GPR35
|
G protein-coupled receptor 35
|
|
chr5_+_31193739
Show fit
|
0.61 |
ENST00000514738.5
|
CDH6
|
cadherin 6
|
|
chr17_+_2796404
Show fit
|
0.60 |
ENST00000366401.8
ENST00000254695.13
ENST00000542807.1
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr18_-_23662868
Show fit
|
0.59 |
ENST00000586087.1
ENST00000592179.6
|
ANKRD29
|
ankyrin repeat domain 29
|
|
chr15_-_74082550
Show fit
|
0.58 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family member A
|
|
chr10_+_86654541
Show fit
|
0.57 |
ENST00000241891.10
ENST00000372071.6
|
OPN4
|
opsin 4
|
|
chr3_-_195811916
Show fit
|
0.56 |
ENST00000463781.8
|
MUC4
|
mucin 4, cell surface associated
|
|
chr9_+_122375286
Show fit
|
0.54 |
ENST00000373698.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chr3_-_195811857
Show fit
|
0.54 |
ENST00000349607.8
ENST00000346145.8
|
MUC4
|
mucin 4, cell surface associated
|
|
chr13_-_52159559
Show fit
|
0.52 |
ENST00000620675.4
ENST00000610828.5
|
NEK3
|
NIMA related kinase 3
|
|
chr3_+_111674654
Show fit
|
0.51 |
ENST00000636933.1
ENST00000393934.7
ENST00000477665.2
|
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2
|
|
chr1_-_169367746
Show fit
|
0.49 |
ENST00000367811.8
ENST00000472647.5
|
NME7
|
NME/NM23 family member 7
|
|
chr4_-_167234552
Show fit
|
0.44 |
ENST00000512648.5
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3
|
|
chr19_-_11155798
Show fit
|
0.44 |
ENST00000592540.5
|
SPC24
|
SPC24 component of NDC80 kinetochore complex
|
|
chr13_-_52159529
Show fit
|
0.43 |
ENST00000618534.4
ENST00000550841.1
|
NEK3
|
NIMA related kinase 3
|
|
chr13_-_52159844
Show fit
|
0.42 |
ENST00000611833.4
|
NEK3
|
NIMA related kinase 3
|
|
chr15_+_75347727
Show fit
|
0.40 |
ENST00000565051.5
ENST00000564257.1
ENST00000567005.1
|
NEIL1
|
nei like DNA glycosylase 1
|
|
chr20_-_38165261
Show fit
|
0.39 |
ENST00000361475.7
|
TGM2
|
transglutaminase 2
|
|
chr12_-_23584600
Show fit
|
0.39 |
ENST00000396007.6
|
SOX5
|
SRY-box transcription factor 5
|
|
chr6_-_75493773
Show fit
|
0.39 |
ENST00000237172.12
|
FILIP1
|
filamin A interacting protein 1
|
|
chr6_-_75493629
Show fit
|
0.38 |
ENST00000393004.6
|
FILIP1
|
filamin A interacting protein 1
|
|
chr4_+_73853290
Show fit
|
0.38 |
ENST00000226524.4
|
PF4V1
|
platelet factor 4 variant 1
|
|
chr15_+_67125707
Show fit
|
0.38 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3
|
|
chr12_-_95996302
Show fit
|
0.37 |
ENST00000261208.8
ENST00000538703.5
ENST00000541929.5
|
HAL
|
histidine ammonia-lyase
|
|
chr19_+_17405685
Show fit
|
0.35 |
ENST00000646744.1
ENST00000635536.2
ENST00000635060.2
ENST00000634291.2
ENST00000645298.1
ENST00000528911.5
|
BISPR
MVB12A
|
BST2 interferon stimulated positive regulator
multivesicular body subunit 12A
|
|
chr4_-_167234266
Show fit
|
0.34 |
ENST00000511269.5
ENST00000506697.5
ENST00000512042.1
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3
|
|
chr17_-_41098084
Show fit
|
0.33 |
ENST00000318329.6
ENST00000333822.5
|
KRTAP4-8
|
keratin associated protein 4-8
|
|
chr1_+_159439722
Show fit
|
0.32 |
ENST00000641630.1
ENST00000423932.6
|
OR10J1
|
olfactory receptor family 10 subfamily J member 1
|
|
chr19_+_17405734
Show fit
|
0.30 |
ENST00000635572.1
ENST00000634675.1
ENST00000634813.1
ENST00000647283.1
ENST00000635435.2
ENST00000634731.2
ENST00000635339.1
ENST00000528604.5
|
BISPR
MVB12A
|
BST2 interferon stimulated positive regulator
multivesicular body subunit 12A
|
|
chr12_-_24562438
Show fit
|
0.30 |
ENST00000646273.1
ENST00000659413.1
ENST00000446891.7
|
SOX5
|
SRY-box transcription factor 5
|
|
chr3_-_116444983
Show fit
|
0.29 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein
|
|
chr22_+_44752552
Show fit
|
0.29 |
ENST00000389774.6
ENST00000356099.11
ENST00000396119.6
ENST00000336963.8
ENST00000412433.5
|
ARHGAP8
|
Rho GTPase activating protein 8
|
|
chrX_-_63351308
Show fit
|
0.28 |
ENST00000374884.3
|
SPIN4
|
spindlin family member 4
|
|
chr7_+_27242700
Show fit
|
0.27 |
ENST00000222761.7
|
EVX1
|
even-skipped homeobox 1
|
|
chr18_+_58864866
Show fit
|
0.27 |
ENST00000588456.5
ENST00000591808.6
ENST00000589481.1
ENST00000591049.1
|
ZNF532
|
zinc finger protein 532
|
|
chr17_-_48722510
Show fit
|
0.26 |
ENST00000290294.5
|
PRAC1
|
PRAC1 small nuclear protein
|
|
chr8_+_26390362
Show fit
|
0.26 |
ENST00000518611.5
|
BNIP3L
|
BCL2 interacting protein 3 like
|
|
chr4_-_167234426
Show fit
|
0.26 |
ENST00000541354.5
ENST00000509854.5
ENST00000512681.5
ENST00000357545.9
ENST00000510741.5
ENST00000510403.5
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3
|
|
chrX_+_17375230
Show fit
|
0.26 |
ENST00000380060.7
|
NHS
|
NHS actin remodeling regulator
|
|
chr19_+_51127030
Show fit
|
0.26 |
ENST00000599948.1
|
SIGLEC9
|
sialic acid binding Ig like lectin 9
|
|
chr4_+_94489030
Show fit
|
0.25 |
ENST00000510099.5
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr4_+_15778320
Show fit
|
0.24 |
ENST00000226279.8
|
CD38
|
CD38 molecule
|
|
chr16_+_6019663
Show fit
|
0.24 |
ENST00000422070.8
|
RBFOX1
|
RNA binding fox-1 homolog 1
|
|
chr15_+_78340344
Show fit
|
0.24 |
ENST00000299529.7
|
CRABP1
|
cellular retinoic acid binding protein 1
|
|
chrX_+_17375194
Show fit
|
0.24 |
ENST00000676302.1
|
NHS
|
NHS actin remodeling regulator
|
|
chr2_-_212124901
Show fit
|
0.24 |
ENST00000402597.6
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4
|
|
chr12_+_65824475
Show fit
|
0.24 |
ENST00000403681.7
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr4_-_167234579
Show fit
|
0.23 |
ENST00000502330.5
ENST00000357154.7
ENST00000421836.6
|
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3
|
|
chr1_+_109910986
Show fit
|
0.23 |
ENST00000369801.1
|
CSF1
|
colony stimulating factor 1
|
|
chr18_+_44680093
Show fit
|
0.23 |
ENST00000426838.8
ENST00000677068.1
|
SETBP1
|
SET binding protein 1
|
|
chr2_+_188291661
Show fit
|
0.23 |
ENST00000409843.5
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1
|
|
chr4_-_151760977
Show fit
|
0.23 |
ENST00000512306.5
ENST00000508611.1
ENST00000515812.5
ENST00000263985.11
|
GATB
|
glutamyl-tRNA amidotransferase subunit B
|
|
chr12_+_132710806
Show fit
|
0.23 |
ENST00000317555.6
|
PGAM5
|
PGAM family member 5, mitochondrial serine/threonine protein phosphatase
|
|
chr14_-_93788475
Show fit
|
0.23 |
ENST00000393140.6
|
PRIMA1
|
proline rich membrane anchor 1
|
|
chr6_-_152563271
Show fit
|
0.23 |
ENST00000535896.7
ENST00000672122.1
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1
|
|
chr19_+_30372364
Show fit
|
0.23 |
ENST00000355537.4
|
ZNF536
|
zinc finger protein 536
|
|
chr8_-_133102874
Show fit
|
0.22 |
ENST00000395352.7
|
SLA
|
Src like adaptor
|
|
chr1_+_169368175
Show fit
|
0.22 |
ENST00000367808.8
ENST00000426663.1
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr1_+_169367934
Show fit
|
0.22 |
ENST00000367807.7
ENST00000329281.6
ENST00000420531.1
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr10_+_88594746
Show fit
|
0.22 |
ENST00000531458.1
|
LIPJ
|
lipase family member J
|
|
chr6_-_30686624
Show fit
|
0.21 |
ENST00000274853.8
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18
|
|
chr3_-_53256009
Show fit
|
0.21 |
ENST00000296289.10
ENST00000462138.6
ENST00000423516.5
|
TKT
|
transketolase
|
|
chr13_-_114132580
Show fit
|
0.21 |
ENST00000334062.8
|
RASA3
|
RAS p21 protein activator 3
|
|
chr5_-_83673544
Show fit
|
0.21 |
ENST00000503117.1
ENST00000510978.5
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr10_+_18260715
Show fit
|
0.20 |
ENST00000615785.4
ENST00000617363.4
ENST00000396576.6
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2
|
|
chr12_+_65278643
Show fit
|
0.20 |
ENST00000355192.8
ENST00000308259.10
ENST00000540804.5
ENST00000535664.5
ENST00000541189.5
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr1_-_228415993
Show fit
|
0.20 |
ENST00000366697.6
|
TRIM17
|
tripartite motif containing 17
|
|
chr1_+_13060769
Show fit
|
0.20 |
ENST00000617807.3
|
HNRNPCL3
|
heterogeneous nuclear ribonucleoprotein C like 3
|
|
chr3_+_14675128
Show fit
|
0.19 |
ENST00000435614.5
ENST00000253697.8
ENST00000412910.1
|
C3orf20
|
chromosome 3 open reading frame 20
|
|
chr2_-_218831791
Show fit
|
0.19 |
ENST00000439262.6
ENST00000430489.1
|
PRKAG3
|
protein kinase AMP-activated non-catalytic subunit gamma 3
|
|
chr5_+_150508110
Show fit
|
0.19 |
ENST00000261797.7
|
NDST1
|
N-deacetylase and N-sulfotransferase 1
|
|
chr16_+_16379055
Show fit
|
0.19 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family member A7
|
|
chr7_+_27242796
Show fit
|
0.19 |
ENST00000496902.7
|
EVX1
|
even-skipped homeobox 1
|
|
chr12_-_47771029
Show fit
|
0.19 |
ENST00000549151.5
ENST00000548919.5
|
RAPGEF3
|
Rap guanine nucleotide exchange factor 3
|
|
chr18_+_34593312
Show fit
|
0.19 |
ENST00000591816.6
ENST00000588125.5
ENST00000684610.1
ENST00000683705.1
ENST00000598334.5
ENST00000588684.5
ENST00000554864.7
ENST00000399121.9
ENST00000595022.5
|
DTNA
|
dystrobrevin alpha
|
|
chr6_-_30687200
Show fit
|
0.18 |
ENST00000399199.7
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18
|
|
chr10_+_18261080
Show fit
|
0.18 |
ENST00000377319.8
ENST00000377331.8
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2
|
|
chr1_+_157993273
Show fit
|
0.17 |
ENST00000360089.8
ENST00000368173.7
|
KIRREL1
|
kirre like nephrin family adhesion molecule 1
|
|
chr3_-_53255990
Show fit
|
0.17 |
ENST00000423525.6
|
TKT
|
transketolase
|
|
chr8_-_133102477
Show fit
|
0.17 |
ENST00000522119.5
ENST00000523610.5
ENST00000338087.10
ENST00000521302.5
ENST00000519558.5
ENST00000519747.5
ENST00000517648.5
|
SLA
|
Src like adaptor
|
|
chr17_+_4715438
Show fit
|
0.17 |
ENST00000571206.1
|
ARRB2
|
arrestin beta 2
|
|
chr12_+_53103479
Show fit
|
0.16 |
ENST00000301466.8
ENST00000551896.5
|
SOAT2
|
sterol O-acyltransferase 2
|
|
chr21_+_44646414
Show fit
|
0.16 |
ENST00000334670.9
|
KRTAP10-11
|
keratin associated protein 10-11
|
|
chr12_-_84911178
Show fit
|
0.15 |
ENST00000681688.1
|
SLC6A15
|
solute carrier family 6 member 15
|
|
chr1_+_15438435
Show fit
|
0.15 |
ENST00000375943.6
ENST00000375949.5
|
CTRC
|
chymotrypsin C
|
|
chr4_+_76435216
Show fit
|
0.15 |
ENST00000296043.7
|
SHROOM3
|
shroom family member 3
|
|
chr8_-_16192772
Show fit
|
0.15 |
ENST00000350896.3
|
MSR1
|
macrophage scavenger receptor 1
|
|
chr1_+_13061158
Show fit
|
0.14 |
ENST00000681473.1
|
HNRNPCL3
|
heterogeneous nuclear ribonucleoprotein C like 3
|
|
chr11_-_18791563
Show fit
|
0.14 |
ENST00000396168.1
|
PTPN5
|
protein tyrosine phosphatase non-receptor type 5
|
|
chr1_+_89364051
Show fit
|
0.14 |
ENST00000370456.5
|
GBP6
|
guanylate binding protein family member 6
|
|
chr22_-_42649332
Show fit
|
0.13 |
ENST00000352397.10
|
CYB5R3
|
cytochrome b5 reductase 3
|
|
chr17_+_60422483
Show fit
|
0.13 |
ENST00000269127.5
|
C17orf64
|
chromosome 17 open reading frame 64
|
|
chr17_+_32021005
Show fit
|
0.13 |
ENST00000327564.11
ENST00000584368.5
ENST00000394713.7
ENST00000341671.11
|
LRRC37B
|
leucine rich repeat containing 37B
|
|
chr16_+_29679132
Show fit
|
0.13 |
ENST00000395384.9
ENST00000562473.1
|
QPRT
|
quinolinate phosphoribosyltransferase
|
|
chr3_+_50246888
Show fit
|
0.13 |
ENST00000451956.1
|
GNAI2
|
G protein subunit alpha i2
|
|
chr12_+_65824403
Show fit
|
0.12 |
ENST00000393578.7
ENST00000425208.6
ENST00000536545.5
ENST00000354636.7
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr15_-_41544243
Show fit
|
0.12 |
ENST00000567866.5
ENST00000304330.9
ENST00000561603.5
ENST00000566863.1
|
RPAP1
|
RNA polymerase II associated protein 1
|
|
chr7_+_64045420
Show fit
|
0.12 |
ENST00000647787.1
ENST00000456806.3
|
ENSG00000285544.1
ZNF727
|
novel transcript
zinc finger protein 727
|
|
chr4_+_6987157
Show fit
|
0.12 |
ENST00000451522.6
|
TBC1D14
|
TBC1 domain family member 14
|
|
chr13_+_30422487
Show fit
|
0.12 |
ENST00000638137.1
ENST00000635918.2
|
UBE2L5
|
ubiquitin conjugating enzyme E2 L5
|
|
chr1_+_231241195
Show fit
|
0.11 |
ENST00000436239.5
ENST00000366647.9
ENST00000416000.1
|
GNPAT
|
glyceronephosphate O-acyltransferase
|
|
chr2_-_189179754
Show fit
|
0.11 |
ENST00000374866.9
ENST00000618828.1
|
COL5A2
|
collagen type V alpha 2 chain
|
|
chr19_-_9538590
Show fit
|
0.11 |
ENST00000593003.5
|
ZNF426
|
zinc finger protein 426
|
|
chr16_+_15643267
Show fit
|
0.10 |
ENST00000396355.5
|
NDE1
|
nudE neurodevelopment protein 1
|
|
chrX_-_72714181
Show fit
|
0.10 |
ENST00000339490.7
ENST00000541944.5
ENST00000373539.3
|
PHKA1
|
phosphorylase kinase regulatory subunit alpha 1
|
|
chr11_+_15114912
Show fit
|
0.10 |
ENST00000379556.8
|
INSC
|
INSC spindle orientation adaptor protein
|
|
chr1_+_115641945
Show fit
|
0.10 |
ENST00000355485.7
ENST00000369510.8
|
VANGL1
|
VANGL planar cell polarity protein 1
|
|
chr1_-_111488795
Show fit
|
0.10 |
ENST00000472933.2
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3
|
|
chr8_-_16192644
Show fit
|
0.10 |
ENST00000262101.10
ENST00000381998.8
|
MSR1
|
macrophage scavenger receptor 1
|
|
chr1_+_209756032
Show fit
|
0.10 |
ENST00000400959.7
ENST00000367025.8
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr15_+_81182579
Show fit
|
0.10 |
ENST00000302987.9
|
IL16
|
interleukin 16
|
|
chr12_+_15911324
Show fit
|
0.09 |
ENST00000524480.5
ENST00000428559.7
ENST00000531803.5
ENST00000532964.5
|
DERA
|
deoxyribose-phosphate aldolase
|
|
chr20_+_43565115
Show fit
|
0.09 |
ENST00000423407.7
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr15_+_63121818
Show fit
|
0.09 |
ENST00000413507.3
|
LACTB
|
lactamase beta
|
|
chr1_+_109910892
Show fit
|
0.09 |
ENST00000369802.7
ENST00000420111.6
|
CSF1
|
colony stimulating factor 1
|
|
chr12_-_6631632
Show fit
|
0.09 |
ENST00000431922.1
|
LPAR5
|
lysophosphatidic acid receptor 5
|
|
chrX_+_141896235
Show fit
|
0.09 |
ENST00000409007.1
|
MAGEC3
|
MAGE family member C3
|
|
chr18_+_34593392
Show fit
|
0.09 |
ENST00000684377.1
|
DTNA
|
dystrobrevin alpha
|
|
chr1_+_226548747
Show fit
|
0.08 |
ENST00000366788.8
ENST00000366789.5
|
STUM
|
stum, mechanosensory transduction mediator homolog
|
|
chr19_+_2360238
Show fit
|
0.08 |
ENST00000649857.1
|
TMPRSS9
|
transmembrane serine protease 9
|
|
chr3_-_116445458
Show fit
|
0.08 |
ENST00000490035.7
|
LSAMP
|
limbic system associated membrane protein
|
|
chr12_+_54549586
Show fit
|
0.08 |
ENST00000243052.8
|
PDE1B
|
phosphodiesterase 1B
|
|
chr5_+_73813518
Show fit
|
0.08 |
ENST00000296799.8
|
ARHGEF28
|
Rho guanine nucleotide exchange factor 28
|
|
chr6_-_34146080
Show fit
|
0.07 |
ENST00000538487.7
ENST00000374181.8
|
GRM4
|
glutamate metabotropic receptor 4
|
|
chrX_+_1268828
Show fit
|
0.07 |
ENST00000432318.8
ENST00000381509.8
ENST00000494969.7
ENST00000355805.7
ENST00000355432.8
|
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha
|
|
chr11_-_4393650
Show fit
|
0.07 |
ENST00000254436.8
|
TRIM21
|
tripartite motif containing 21
|
|
chr10_-_48605032
Show fit
|
0.07 |
ENST00000249601.9
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr11_+_111937320
Show fit
|
0.07 |
ENST00000440460.7
|
DIXDC1
|
DIX domain containing 1
|
|
chr11_+_10455292
Show fit
|
0.07 |
ENST00000396553.6
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr6_+_29827817
Show fit
|
0.07 |
ENST00000360323.11
ENST00000376818.7
ENST00000376815.3
|
HLA-G
|
major histocompatibility complex, class I, G
|
|
chr6_+_30720335
Show fit
|
0.07 |
ENST00000327892.13
|
TUBB
|
tubulin beta class I
|
|
chr12_+_6924449
Show fit
|
0.07 |
ENST00000356654.8
|
ATN1
|
atrophin 1
|
|
chr7_-_78771265
Show fit
|
0.06 |
ENST00000630991.2
ENST00000629359.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr7_-_50565381
Show fit
|
0.06 |
ENST00000444124.7
|
DDC
|
dopa decarboxylase
|
|
chr14_+_24429932
Show fit
|
0.06 |
ENST00000556842.5
ENST00000553935.6
|
KHNYN
|
KH and NYN domain containing
|
|
chr15_+_63121852
Show fit
|
0.06 |
ENST00000261893.9
|
LACTB
|
lactamase beta
|
|
chr12_+_120650492
Show fit
|
0.06 |
ENST00000351200.6
|
CABP1
|
calcium binding protein 1
|
|
chr12_-_10998304
Show fit
|
0.06 |
ENST00000538986.2
|
TAS2R20
|
taste 2 receptor member 20
|
|
chrX_+_120362079
Show fit
|
0.05 |
ENST00000539306.5
ENST00000218008.8
ENST00000361319.3
|
ATP1B4
|
ATPase Na+/K+ transporting family member beta 4
|
|
chr13_+_60396946
Show fit
|
0.05 |
ENST00000377894.6
|
TDRD3
|
tudor domain containing 3
|
|
chr13_+_111114550
Show fit
|
0.05 |
ENST00000317133.9
|
ARHGEF7
|
Rho guanine nucleotide exchange factor 7
|
|
chr15_-_62165274
Show fit
|
0.05 |
ENST00000380392.4
|
C2CD4B
|
C2 calcium dependent domain containing 4B
|
|
chr17_-_58328756
Show fit
|
0.05 |
ENST00000268893.10
ENST00000343736.9
|
TSPOAP1
|
TSPO associated protein 1
|
|
chr7_-_135510074
Show fit
|
0.05 |
ENST00000428680.6
ENST00000423368.6
ENST00000451834.5
ENST00000361528.8
ENST00000541284.5
ENST00000315544.6
|
CNOT4
|
CCR4-NOT transcription complex subunit 4
|
|
chr17_-_7178116
Show fit
|
0.05 |
ENST00000380920.8
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr12_+_10010627
Show fit
|
0.05 |
ENST00000338896.11
ENST00000396502.5
|
CLEC12B
|
C-type lectin domain family 12 member B
|
|
chr19_-_42132465
Show fit
|
0.05 |
ENST00000529067.5
ENST00000529952.5
ENST00000342301.8
ENST00000389341.9
|
POU2F2
|
POU class 2 homeobox 2
|
|
chr8_+_43056409
Show fit
|
0.04 |
ENST00000525699.5
ENST00000529687.5
|
FNTA
|
farnesyltransferase, CAAX box, alpha
|
|
chr19_-_38224215
Show fit
|
0.04 |
ENST00000355526.10
ENST00000420980.7
ENST00000614244.4
|
DPF1
|
double PHD fingers 1
|
|
chr3_+_130850585
Show fit
|
0.04 |
ENST00000505330.5
ENST00000504381.5
ENST00000507488.6
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1
|
|
chr16_+_70294777
Show fit
|
0.04 |
ENST00000563206.5
ENST00000451014.7
ENST00000568625.5
|
DDX19B
|
DEAD-box helicase 19B
|
|
chr13_+_60397214
Show fit
|
0.04 |
ENST00000377881.8
|
TDRD3
|
tudor domain containing 3
|
|
chr1_+_209756149
Show fit
|
0.04 |
ENST00000367026.7
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chrX_-_142205260
Show fit
|
0.04 |
ENST00000247452.4
|
MAGEC2
|
MAGE family member C2
|
|
chr11_+_61950370
Show fit
|
0.04 |
ENST00000449131.6
|
BEST1
|
bestrophin 1
|
|
chr9_-_111328520
Show fit
|
0.03 |
ENST00000374428.1
|
OR2K2
|
olfactory receptor family 2 subfamily K member 2
|
|
chr14_+_52707178
Show fit
|
0.03 |
ENST00000612399.4
|
PSMC6
|
proteasome 26S subunit, ATPase 6
|
|
chr11_+_111879482
Show fit
|
0.03 |
ENST00000260276.8
ENST00000530214.5
ENST00000530799.5
|
C11orf1
|
chromosome 11 open reading frame 1
|
|
chr19_+_47332167
Show fit
|
0.03 |
ENST00000595464.3
|
C5AR2
|
complement component 5a receptor 2
|
|
chrX_+_1268807
Show fit
|
0.03 |
ENST00000381524.8
ENST00000381529.9
ENST00000412290.6
|
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha
|
|
chr22_+_44026283
Show fit
|
0.03 |
ENST00000619710.4
|
PARVB
|
parvin beta
|
|
chr14_+_21030201
Show fit
|
0.03 |
ENST00000321760.11
ENST00000460647.6
ENST00000530140.6
ENST00000472458.5
|
TPPP2
|
tubulin polymerization promoting protein family member 2
|
|
chr1_-_153616289
Show fit
|
0.03 |
ENST00000368701.5
ENST00000344616.4
|
S100A14
|
S100 calcium binding protein A14
|
|
chr19_-_3801791
Show fit
|
0.03 |
ENST00000590849.1
ENST00000395045.6
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr14_+_52707192
Show fit
|
0.03 |
ENST00000445930.7
ENST00000555339.5
ENST00000556813.1
|
PSMC6
|
proteasome 26S subunit, ATPase 6
|
|
chr19_-_42132391
Show fit
|
0.03 |
ENST00000528894.8
ENST00000560804.6
ENST00000560558.5
ENST00000560398.5
ENST00000526816.6
ENST00000625670.2
|
POU2F2
|
POU class 2 homeobox 2
|
|
chr6_-_145735964
Show fit
|
0.02 |
ENST00000640980.1
ENST00000639423.1
ENST00000611340.5
|
EPM2A
|
EPM2A glucan phosphatase, laforin
|
|
chr6_-_15548360
Show fit
|
0.02 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1
|
|
chrX_+_1268786
Show fit
|
0.02 |
ENST00000501036.7
ENST00000417535.7
|
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha
|
|
chr13_-_79406175
Show fit
|
0.02 |
ENST00000438724.5
ENST00000622611.4
ENST00000438737.3
|
RBM26
|
RNA binding motif protein 26
|
|
chr4_-_141133436
Show fit
|
0.02 |
ENST00000306799.7
ENST00000515673.7
|
RNF150
|
ring finger protein 150
|
|
chr12_-_11134644
Show fit
|
0.02 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30
|
|
chrY_+_9337464
Show fit
|
0.02 |
ENST00000426950.6
ENST00000640033.1
ENST00000383008.1
|
TSPY4
|
testis specific protein Y-linked 4
|
|
chr19_-_35135180
Show fit
|
0.02 |
ENST00000392225.7
|
LGI4
|
leucine rich repeat LGI family member 4
|
|
chr11_+_61950063
Show fit
|
0.02 |
ENST00000534553.5
|
BEST1
|
bestrophin 1
|
|
chr15_+_72654738
Show fit
|
0.02 |
ENST00000421285.3
|
GOLGA6B
|
golgin A6 family member B
|
|
chr19_+_571274
Show fit
|
0.02 |
ENST00000545507.6
ENST00000346916.9
|
BSG
|
basigin (Ok blood group)
|
|
chr3_+_184276007
Show fit
|
0.02 |
ENST00000359140.8
ENST00000404464.8
ENST00000357474.9
|
ECE2
|
endothelin converting enzyme 2
|
|
chr7_-_149126306
Show fit
|
0.02 |
ENST00000483014.1
ENST00000378061.7
|
ZNF425
|
zinc finger protein 425
|
|
chrY_+_9398421
Show fit
|
0.01 |
ENST00000457222.6
ENST00000440483.4
ENST00000424594.1
|
TSPY3
|
testis specific protein Y-linked 3
|
|
chr2_-_99336306
Show fit
|
0.01 |
ENST00000264255.8
ENST00000409434.5
ENST00000434323.5
|
TXNDC9
|
thioredoxin domain containing 9
|
|
chrY_+_9527880
Show fit
|
0.01 |
ENST00000428845.6
ENST00000444056.1
|
TSPY10
|
testis specific protein Y-linked 10
|
|
chr5_+_153490727
Show fit
|
0.01 |
ENST00000340592.9
|
GRIA1
|
glutamate ionotropic receptor AMPA type subunit 1
|
|
chr16_+_4624811
Show fit
|
0.01 |
ENST00000415496.5
ENST00000262370.12
ENST00000587747.5
ENST00000399577.9
ENST00000588994.5
ENST00000586183.5
|
MGRN1
|
mahogunin ring finger 1
|
|
chr22_-_37149900
Show fit
|
0.01 |
ENST00000216223.10
|
IL2RB
|
interleukin 2 receptor subunit beta
|
|
chr14_+_23185316
Show fit
|
0.01 |
ENST00000399910.5
ENST00000492621.5
|
RNF212B
|
ring finger protein 212B
|
|
chr19_+_7595830
Show fit
|
0.01 |
ENST00000160298.9
ENST00000446248.4
|
CAMSAP3
|
calmodulin regulated spectrin associated protein family member 3
|