Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
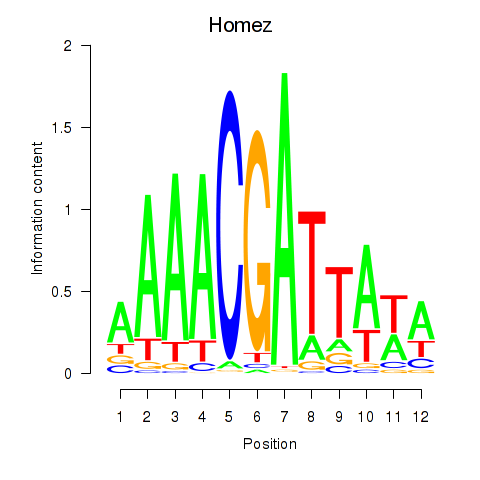
Results for Homez
Z-value: 0.92
Transcription factors associated with Homez
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Homez
|
ENSMUSG00000057156.9 | homeodomain leucine zipper-encoding gene |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Homez | mm10_v2_chr14_-_54864055_54864158 | -0.46 | 4.7e-03 | Click! |
Activity profile of Homez motif
Sorted Z-values of Homez motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_8405060 | 3.78 |
ENSMUST00000064507.5
ENSMUST00000120540.1 ENSMUST00000096269.4 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr4_-_61835185 | 3.27 |
ENSMUST00000082287.2
|
Mup5
|
major urinary protein 5 |
| chr8_-_93229517 | 3.19 |
ENSMUST00000176282.1
ENSMUST00000034173.7 |
Ces1e
|
carboxylesterase 1E |
| chr2_-_24049389 | 3.13 |
ENSMUST00000051416.5
|
Hnmt
|
histamine N-methyltransferase |
| chr6_-_55175019 | 2.42 |
ENSMUST00000003569.5
|
Inmt
|
indolethylamine N-methyltransferase |
| chr6_+_122006798 | 2.18 |
ENSMUST00000081777.6
|
Mug2
|
murinoglobulin 2 |
| chr17_+_79626669 | 2.09 |
ENSMUST00000086570.1
|
4921513D11Rik
|
RIKEN cDNA 4921513D11 gene |
| chr13_-_8871751 | 1.71 |
ENSMUST00000175958.1
|
Wdr37
|
WD repeat domain 37 |
| chr3_+_138415484 | 1.54 |
ENSMUST00000161312.1
ENSMUST00000013458.8 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr6_+_149141638 | 1.50 |
ENSMUST00000166416.1
ENSMUST00000111551.1 |
Mettl20
|
methyltransferase like 20 |
| chr11_-_74925658 | 1.49 |
ENSMUST00000138612.1
ENSMUST00000123855.1 ENSMUST00000128556.1 ENSMUST00000108448.1 ENSMUST00000108447.1 ENSMUST00000065211.2 |
Srr
|
serine racemase |
| chr8_+_46010596 | 1.42 |
ENSMUST00000110381.2
|
Lrp2bp
|
Lrp2 binding protein |
| chr15_-_5063741 | 1.38 |
ENSMUST00000110689.3
|
C7
|
complement component 7 |
| chr18_-_3309858 | 1.32 |
ENSMUST00000144496.1
ENSMUST00000154715.1 |
Crem
|
cAMP responsive element modulator |
| chr1_+_78657874 | 1.31 |
ENSMUST00000134566.1
ENSMUST00000142704.1 ENSMUST00000053760.5 |
Acsl3
Utp14b
|
acyl-CoA synthetase long-chain family member 3 UTP14, U3 small nucleolar ribonucleoprotein, homolog B (yeast) |
| chr1_-_72212249 | 1.31 |
ENSMUST00000048860.7
|
Mreg
|
melanoregulin |
| chr1_+_13668739 | 1.30 |
ENSMUST00000088542.3
|
Xkr9
|
X Kell blood group precursor related family member 9 homolog |
| chr16_+_11406618 | 1.27 |
ENSMUST00000122168.1
|
Snx29
|
sorting nexin 29 |
| chr1_+_78657825 | 1.21 |
ENSMUST00000035779.8
|
Acsl3
|
acyl-CoA synthetase long-chain family member 3 |
| chr10_+_88459569 | 1.20 |
ENSMUST00000020252.3
ENSMUST00000125612.1 |
Sycp3
|
synaptonemal complex protein 3 |
| chr2_-_3419066 | 1.18 |
ENSMUST00000115082.3
|
Meig1
|
meiosis expressed gene 1 |
| chr10_-_24101951 | 1.17 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr9_-_78322357 | 1.17 |
ENSMUST00000095071.4
|
Gm8074
|
predicted gene 8074 |
| chr16_+_91269759 | 1.16 |
ENSMUST00000056882.5
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr13_-_8871696 | 1.15 |
ENSMUST00000054251.6
ENSMUST00000176813.1 |
Wdr37
|
WD repeat domain 37 |
| chr12_+_103434211 | 1.12 |
ENSMUST00000079294.5
ENSMUST00000076788.5 ENSMUST00000076702.5 ENSMUST00000066701.6 ENSMUST00000085065.5 ENSMUST00000140838.1 |
Ifi27
|
interferon, alpha-inducible protein 27 |
| chr10_-_18234930 | 1.12 |
ENSMUST00000052648.8
ENSMUST00000080860.6 ENSMUST00000173243.1 |
Ccdc28a
|
coiled-coil domain containing 28A |
| chr7_+_27119909 | 1.08 |
ENSMUST00000003100.8
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2 |
| chr5_+_119625939 | 1.06 |
ENSMUST00000156235.1
|
Gm16063
|
predicted gene 16063 |
| chr7_+_49910112 | 1.05 |
ENSMUST00000056442.5
|
Slc6a5
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 5 |
| chr1_+_169969409 | 1.00 |
ENSMUST00000180638.1
|
3110045C21Rik
|
RIKEN cDNA 3110045C21 gene |
| chr17_+_21491256 | 0.99 |
ENSMUST00000076664.6
|
Zfp53
|
zinc finger protein 53 |
| chr15_-_98221056 | 0.99 |
ENSMUST00000170618.1
ENSMUST00000141911.1 |
Olfr287
|
olfactory receptor 287 |
| chr3_+_85915722 | 0.97 |
ENSMUST00000054148.7
|
Gm9790
|
predicted gene 9790 |
| chr2_-_3419019 | 0.97 |
ENSMUST00000115084.1
ENSMUST00000115083.1 |
Meig1
|
meiosis expressed gene 1 |
| chr12_-_99393010 | 0.97 |
ENSMUST00000177451.1
|
Foxn3
|
forkhead box N3 |
| chr1_+_4857760 | 0.95 |
ENSMUST00000081551.7
|
Tcea1
|
transcription elongation factor A (SII) 1 |
| chr9_+_108339048 | 0.94 |
ENSMUST00000082429.5
|
Gpx1
|
glutathione peroxidase 1 |
| chr10_-_115362191 | 0.94 |
ENSMUST00000092170.5
|
Tmem19
|
transmembrane protein 19 |
| chr18_+_84935158 | 0.93 |
ENSMUST00000037718.9
|
Fbxo15
|
F-box protein 15 |
| chr16_+_43235856 | 0.92 |
ENSMUST00000146708.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr17_-_57059795 | 0.89 |
ENSMUST00000040280.7
|
Slc25a23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr11_-_40695203 | 0.87 |
ENSMUST00000101347.3
|
Mat2b
|
methionine adenosyltransferase II, beta |
| chr15_+_90224293 | 0.85 |
ENSMUST00000100309.1
|
Alg10b
|
asparagine-linked glycosylation 10B (alpha-1,2-glucosyltransferase) |
| chrX_+_36795642 | 0.85 |
ENSMUST00000016463.3
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr15_+_100353149 | 0.84 |
ENSMUST00000075675.5
ENSMUST00000088142.5 ENSMUST00000176287.1 |
AB099516
Mettl7a2
|
cDNA sequence AB099516 methyltransferase like 7A2 |
| chr6_+_137754529 | 0.84 |
ENSMUST00000087675.6
|
Dera
|
2-deoxyribose-5-phosphate aldolase homolog (C. elegans) |
| chr10_-_81291227 | 0.83 |
ENSMUST00000045744.6
|
Tjp3
|
tight junction protein 3 |
| chr5_-_113989739 | 0.82 |
ENSMUST00000077689.7
|
Ssh1
|
slingshot homolog 1 (Drosophila) |
| chr3_-_90514250 | 0.80 |
ENSMUST00000107340.1
ENSMUST00000060738.8 |
S100a1
|
S100 calcium binding protein A1 |
| chr5_-_45450143 | 0.80 |
ENSMUST00000154962.1
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr13_-_92131494 | 0.79 |
ENSMUST00000099326.3
ENSMUST00000146492.1 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr3_-_79841729 | 0.78 |
ENSMUST00000168038.1
|
Tmem144
|
transmembrane protein 144 |
| chr2_+_122765237 | 0.77 |
ENSMUST00000005953.4
|
Sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr2_-_127792467 | 0.75 |
ENSMUST00000135091.1
|
1500011K16Rik
|
RIKEN cDNA 1500011K16 gene |
| chr2_-_58052832 | 0.74 |
ENSMUST00000090940.5
|
Ermn
|
ermin, ERM-like protein |
| chr12_-_48559971 | 0.72 |
ENSMUST00000169406.1
|
Gm1818
|
predicted gene 1818 |
| chr8_+_70673364 | 0.72 |
ENSMUST00000146972.1
|
Lsm4
|
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr6_+_58640536 | 0.72 |
ENSMUST00000145161.1
ENSMUST00000114294.1 |
Abcg2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr18_-_3309723 | 0.71 |
ENSMUST00000136961.1
ENSMUST00000152108.1 |
Crem
|
cAMP responsive element modulator |
| chr8_+_21287409 | 0.71 |
ENSMUST00000098893.3
|
Defa3
|
defensin, alpha, 3 |
| chr15_+_9071252 | 0.71 |
ENSMUST00000100789.4
ENSMUST00000100790.3 ENSMUST00000067760.4 |
Nadk2
|
NAD kinase 2, mitochondrial |
| chr8_-_70506710 | 0.71 |
ENSMUST00000136913.1
ENSMUST00000075175.5 |
2810428I15Rik
|
RIKEN cDNA 2810428I15 gene |
| chr5_+_3845171 | 0.70 |
ENSMUST00000044039.1
ENSMUST00000143027.1 |
Lrrd1
|
leucine rich repeats and death domain containing 1 |
| chr13_+_70882948 | 0.70 |
ENSMUST00000022091.3
|
8030423J24Rik
|
RIKEN cDNA 8030423J24 gene |
| chr9_+_55150050 | 0.70 |
ENSMUST00000122441.1
|
Ube2q2
|
ubiquitin-conjugating enzyme E2Q (putative) 2 |
| chr7_-_38227975 | 0.70 |
ENSMUST00000098513.4
|
Plekhf1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr5_-_149636164 | 0.69 |
ENSMUST00000076410.4
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr15_+_5185700 | 0.68 |
ENSMUST00000081640.5
|
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr1_-_136953600 | 0.67 |
ENSMUST00000168126.1
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr16_+_48073444 | 0.66 |
ENSMUST00000088296.2
|
Gm7275
|
predicted gene 7275 |
| chr6_+_71282280 | 0.65 |
ENSMUST00000080949.7
|
Krcc1
|
lysine-rich coiled-coil 1 |
| chr16_-_91618986 | 0.65 |
ENSMUST00000143058.1
ENSMUST00000049244.8 ENSMUST00000169982.1 ENSMUST00000133731.1 |
Dnajc28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr5_-_21785115 | 0.65 |
ENSMUST00000115193.1
ENSMUST00000115192.1 ENSMUST00000115195.1 ENSMUST00000030771.5 |
Dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr9_+_22454290 | 0.64 |
ENSMUST00000168332.1
|
Gm17545
|
predicted gene, 17545 |
| chr12_-_54695813 | 0.64 |
ENSMUST00000110713.3
|
Eapp
|
E2F-associated phosphoprotein |
| chr16_+_11405648 | 0.63 |
ENSMUST00000096273.2
|
Snx29
|
sorting nexin 29 |
| chr1_-_175979114 | 0.63 |
ENSMUST00000104983.1
|
B020018G12Rik
|
RIKEN cDNA B020018G12 gene |
| chr3_+_127791374 | 0.61 |
ENSMUST00000171621.1
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr9_-_78587968 | 0.60 |
ENSMUST00000117645.1
ENSMUST00000119213.1 ENSMUST00000052441.5 |
Slc17a5
|
solute carrier family 17 (anion/sugar transporter), member 5 |
| chrY_-_35920097 | 0.60 |
ENSMUST00000180332.1
|
Gm20896
|
predicted gene, 20896 |
| chr15_+_79659196 | 0.60 |
ENSMUST00000023064.7
|
Cby1
|
chibby homolog 1 (Drosophila) |
| chr6_+_128662379 | 0.59 |
ENSMUST00000032518.4
|
Clec2h
|
C-type lectin domain family 2, member h |
| chr2_+_85975213 | 0.59 |
ENSMUST00000082191.2
|
Olfr1029
|
olfactory receptor 1029 |
| chr5_+_96173940 | 0.58 |
ENSMUST00000048361.8
|
Gm2840
|
predicted gene 2840 |
| chr13_-_85127514 | 0.58 |
ENSMUST00000179230.1
|
Gm4076
|
predicted gene 4076 |
| chr18_-_66002612 | 0.58 |
ENSMUST00000120461.1
ENSMUST00000048260.7 |
Lman1
|
lectin, mannose-binding, 1 |
| chr10_-_41709297 | 0.58 |
ENSMUST00000019955.9
ENSMUST00000099932.3 |
Ccdc162
|
coiled-coil domain containing 162 |
| chr11_+_101155884 | 0.58 |
ENSMUST00000043654.9
|
Tubg2
|
tubulin, gamma 2 |
| chr8_+_21655780 | 0.57 |
ENSMUST00000079528.5
|
Defa17
|
defensin, alpha, 17 |
| chr11_+_101627942 | 0.57 |
ENSMUST00000010506.3
|
Rdm1
|
RAD52 motif 1 |
| chr5_-_66004278 | 0.57 |
ENSMUST00000067737.5
|
9130230L23Rik
|
RIKEN cDNA 9130230L23 gene |
| chr1_+_44147847 | 0.56 |
ENSMUST00000027214.3
|
Ercc5
|
excision repair cross-complementing rodent repair deficiency, complementation group 5 |
| chr8_-_116921408 | 0.56 |
ENSMUST00000078589.6
ENSMUST00000148235.1 |
Cmc2
|
COX assembly mitochondrial protein 2 |
| chr11_-_69369377 | 0.55 |
ENSMUST00000092971.6
ENSMUST00000108661.1 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr18_+_35770318 | 0.55 |
ENSMUST00000165299.1
|
Gm16490
|
predicted gene 16490 |
| chr1_+_96872221 | 0.55 |
ENSMUST00000181489.1
|
Gm5101
|
predicted gene 5101 |
| chr3_+_138143429 | 0.54 |
ENSMUST00000040321.6
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr13_-_76018524 | 0.54 |
ENSMUST00000050997.1
ENSMUST00000179078.1 ENSMUST00000167271.1 |
Rfesd
|
Rieske (Fe-S) domain containing |
| chr17_-_33713372 | 0.54 |
ENSMUST00000173392.1
|
March2
|
membrane-associated ring finger (C3HC4) 2 |
| chr15_+_5185519 | 0.54 |
ENSMUST00000118193.1
ENSMUST00000022751.8 |
Ttc33
|
tetratricopeptide repeat domain 33 |
| chr11_-_96916407 | 0.54 |
ENSMUST00000130774.1
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr11_-_96916448 | 0.53 |
ENSMUST00000103152.4
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr5_+_8422908 | 0.53 |
ENSMUST00000170496.1
|
Slc25a40
|
solute carrier family 25, member 40 |
| chr6_+_88724412 | 0.53 |
ENSMUST00000113585.2
|
Mgll
|
monoglyceride lipase |
| chr5_-_87490869 | 0.52 |
ENSMUST00000147854.1
|
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr10_+_34483400 | 0.52 |
ENSMUST00000019913.7
ENSMUST00000170771.1 |
Frk
|
fyn-related kinase |
| chr17_-_22007301 | 0.52 |
ENSMUST00000075018.3
|
Gm9772
|
predicted gene 9772 |
| chr1_-_59003443 | 0.52 |
ENSMUST00000054653.6
|
Als2cr11
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11 (human) |
| chr8_-_116921365 | 0.51 |
ENSMUST00000128304.1
|
Cmc2
|
COX assembly mitochondrial protein 2 |
| chr2_-_30359278 | 0.51 |
ENSMUST00000163668.2
ENSMUST00000028214.8 ENSMUST00000113621.3 |
Sh3glb2
|
SH3-domain GRB2-like endophilin B2 |
| chr1_-_13127163 | 0.51 |
ENSMUST00000047577.6
|
Prdm14
|
PR domain containing 14 |
| chr14_+_56887795 | 0.51 |
ENSMUST00000022511.8
|
Zmym2
|
zinc finger, MYM-type 2 |
| chr11_-_48826500 | 0.50 |
ENSMUST00000161192.2
|
Gm12184
|
predicted gene 12184 |
| chrM_+_7759 | 0.50 |
ENSMUST00000082407.1
ENSMUST00000082408.1 |
mt-Atp8
mt-Atp6
|
mitochondrially encoded ATP synthase 8 mitochondrially encoded ATP synthase 6 |
| chr4_+_148602527 | 0.50 |
ENSMUST00000105701.2
ENSMUST00000052060.6 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr15_-_83464595 | 0.50 |
ENSMUST00000171436.1
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr2_-_148038270 | 0.50 |
ENSMUST00000132070.1
|
9030622O22Rik
|
RIKEN cDNA 9030622O22 gene |
| chr1_+_163994866 | 0.50 |
ENSMUST00000111490.1
ENSMUST00000045694.7 |
Mettl18
|
methyltransferase like 18 |
| chr17_+_26542760 | 0.49 |
ENSMUST00000090257.4
|
Gm8225
|
predicted gene 8225 |
| chr11_+_29526423 | 0.48 |
ENSMUST00000136351.1
ENSMUST00000020749.6 ENSMUST00000144321.1 ENSMUST00000093239.4 |
Mtif2
|
mitochondrial translational initiation factor 2 |
| chr7_+_143475094 | 0.48 |
ENSMUST00000105917.2
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr8_-_46080284 | 0.48 |
ENSMUST00000177186.1
|
Snx25
|
sorting nexin 25 |
| chr19_-_39649046 | 0.48 |
ENSMUST00000067328.6
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr8_-_129065488 | 0.48 |
ENSMUST00000125112.1
ENSMUST00000108747.2 ENSMUST00000095158.4 |
Ccdc7
|
coiled-coil domain containing 7 |
| chr13_-_92483996 | 0.47 |
ENSMUST00000040106.7
|
Fam151b
|
family with sequence similarity 151, member B |
| chr17_+_21093172 | 0.47 |
ENSMUST00000169389.1
|
Gm6811
|
predicted gene 6811 |
| chr4_-_14826582 | 0.47 |
ENSMUST00000117268.1
|
Otud6b
|
OTU domain containing 6B |
| chr7_-_104917294 | 0.47 |
ENSMUST00000071362.2
|
Olfr667
|
olfactory receptor 667 |
| chr19_-_12501996 | 0.46 |
ENSMUST00000045521.7
|
Dtx4
|
deltex 4 homolog (Drosophila) |
| chr13_+_4049001 | 0.45 |
ENSMUST00000118717.2
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr5_-_149636331 | 0.45 |
ENSMUST00000074846.7
ENSMUST00000110498.1 ENSMUST00000127977.1 ENSMUST00000132412.1 |
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chrX_-_7899220 | 0.45 |
ENSMUST00000033497.2
|
Pqbp1
|
polyglutamine binding protein 1 |
| chr8_-_104534630 | 0.45 |
ENSMUST00000162466.1
ENSMUST00000034349.9 |
Nae1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr1_-_181144133 | 0.45 |
ENSMUST00000027797.7
|
Nvl
|
nuclear VCP-like |
| chr10_-_41587753 | 0.45 |
ENSMUST00000160751.1
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr7_+_120173847 | 0.44 |
ENSMUST00000033201.5
|
Anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr18_-_64516547 | 0.44 |
ENSMUST00000025483.9
|
Nars
|
asparaginyl-tRNA synthetase |
| chr16_+_72663143 | 0.44 |
ENSMUST00000023600.7
|
Robo1
|
roundabout homolog 1 (Drosophila) |
| chr8_-_21327015 | 0.44 |
ENSMUST00000098890.3
|
Defa-rs1
|
defensin, alpha, related sequence 1 |
| chr3_+_5214763 | 0.44 |
ENSMUST00000177924.1
|
Gm10748
|
predicted gene 10748 |
| chr7_-_46958475 | 0.43 |
ENSMUST00000094398.4
|
Uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr5_-_24581879 | 0.43 |
ENSMUST00000088299.4
|
Gm10221
|
predicted gene 10221 |
| chr14_-_34522801 | 0.42 |
ENSMUST00000022325.1
|
9230112D13Rik
|
RIKEN cDNA 9230112D13 gene |
| chr11_-_76763550 | 0.42 |
ENSMUST00000010536.8
|
Gosr1
|
golgi SNAP receptor complex member 1 |
| chr7_+_44676965 | 0.42 |
ENSMUST00000094460.1
|
2310016G11Rik
|
RIKEN cDNA 2310016G11 gene |
| chr11_-_14599194 | 0.42 |
ENSMUST00000139973.1
|
1700046C09Rik
|
RIKEN cDNA 1700046C09 gene |
| chr11_-_62281342 | 0.41 |
ENSMUST00000072916.4
|
Zswim7
|
zinc finger SWIM-type containing 7 |
| chr1_-_36244245 | 0.41 |
ENSMUST00000046875.7
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr8_+_21055047 | 0.41 |
ENSMUST00000098899.3
|
Defa23
|
defensin, alpha, 23 |
| chr13_+_22293193 | 0.40 |
ENSMUST00000091735.1
|
Vmn1r196
|
vomeronasal 1 receptor 196 |
| chrX_-_7899196 | 0.40 |
ENSMUST00000115654.1
|
Pqbp1
|
polyglutamine binding protein 1 |
| chrX_-_37878944 | 0.40 |
ENSMUST00000071885.6
|
Rhox8
|
reproductive homeobox 8 |
| chr5_+_21785253 | 0.40 |
ENSMUST00000030769.5
|
Psmc2
|
proteasome (prosome, macropain) 26S subunit, ATPase 2 |
| chr9_+_89199319 | 0.39 |
ENSMUST00000138109.1
|
Mthfs
|
5, 10-methenyltetrahydrofolate synthetase |
| chr16_-_18876655 | 0.39 |
ENSMUST00000023391.8
|
Mrpl40
|
mitochondrial ribosomal protein L40 |
| chr7_+_45514581 | 0.39 |
ENSMUST00000151506.1
ENSMUST00000085331.5 ENSMUST00000126061.1 |
Tulp2
|
tubby-like protein 2 |
| chr5_-_110108154 | 0.39 |
ENSMUST00000077220.7
|
Gtpbp6
|
GTP binding protein 6 (putative) |
| chr18_-_11051479 | 0.39 |
ENSMUST00000180789.1
|
1010001N08Rik
|
RIKEN cDNA 1010001N08 gene |
| chr7_-_48643801 | 0.38 |
ENSMUST00000094383.1
|
Mrgprb3
|
MAS-related GPR, member B3 |
| chr1_-_138619687 | 0.38 |
ENSMUST00000027642.2
|
Nek7
|
NIMA (never in mitosis gene a)-related expressed kinase 7 |
| chr10_-_89732253 | 0.38 |
ENSMUST00000020109.3
|
Actr6
|
ARP6 actin-related protein 6 |
| chr5_-_120812484 | 0.38 |
ENSMUST00000125547.1
|
Oas1c
|
2'-5' oligoadenylate synthetase 1C |
| chr13_-_19395728 | 0.37 |
ENSMUST00000039694.7
|
Stard3nl
|
STARD3 N-terminal like |
| chr18_-_43477764 | 0.37 |
ENSMUST00000057110.9
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr2_-_30359190 | 0.37 |
ENSMUST00000100215.4
ENSMUST00000113620.3 |
Sh3glb2
|
SH3-domain GRB2-like endophilin B2 |
| chr3_+_157534103 | 0.37 |
ENSMUST00000106058.1
|
Zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr6_-_128581597 | 0.37 |
ENSMUST00000060574.7
|
BC048546
|
cDNA sequence BC048546 |
| chr6_+_142413441 | 0.36 |
ENSMUST00000088263.4
|
B230216G23Rik
|
RIKEN cDNA B230216G23 gene |
| chr2_+_61711694 | 0.36 |
ENSMUST00000028278.7
|
Psmd14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr12_-_69184056 | 0.36 |
ENSMUST00000054544.6
|
Rpl36al
|
ribosomal protein L36A-like |
| chr19_-_30175414 | 0.36 |
ENSMUST00000025778.7
|
Gldc
|
glycine decarboxylase |
| chr5_+_129846980 | 0.36 |
ENSMUST00000171300.1
|
Sumf2
|
sulfatase modifying factor 2 |
| chr16_-_49297394 | 0.35 |
ENSMUST00000080694.2
|
Gm5407
|
predicted gene 5407 |
| chr6_+_37530173 | 0.35 |
ENSMUST00000040987.7
|
Akr1d1
|
aldo-keto reductase family 1, member D1 |
| chr19_+_47731743 | 0.35 |
ENSMUST00000099353.4
|
Sfr1
|
SWI5 dependent recombination repair 1 |
| chr7_-_30195046 | 0.35 |
ENSMUST00000001845.5
|
Capns1
|
calpain, small subunit 1 |
| chr1_+_93342854 | 0.35 |
ENSMUST00000027494.4
|
Ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr2_-_156004147 | 0.35 |
ENSMUST00000156993.1
ENSMUST00000141437.1 |
6430550D23Rik
|
RIKEN cDNA 6430550D23 gene |
| chr3_+_100162381 | 0.35 |
ENSMUST00000029459.3
ENSMUST00000106997.1 |
Gdap2
|
ganglioside-induced differentiation-associated-protein 2 |
| chr4_-_116994374 | 0.34 |
ENSMUST00000030446.8
|
Urod
|
uroporphyrinogen decarboxylase |
| chr17_+_56990264 | 0.34 |
ENSMUST00000002735.7
|
Clpp
|
ClpP caseinolytic peptidase, ATP-dependent, proteolytic subunit |
| chr12_-_69183986 | 0.34 |
ENSMUST00000110620.1
ENSMUST00000110619.1 |
Rpl36al
|
ribosomal protein L36A-like |
| chr5_+_114923234 | 0.34 |
ENSMUST00000031540.4
ENSMUST00000112143.3 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr5_+_87666200 | 0.34 |
ENSMUST00000094641.4
|
Csn1s1
|
casein alpha s1 |
| chr8_+_88199194 | 0.34 |
ENSMUST00000119033.1
ENSMUST00000066748.3 ENSMUST00000118952.1 |
Papd5
|
PAP associated domain containing 5 |
| chr14_-_20393473 | 0.34 |
ENSMUST00000061444.3
|
Mrps16
|
mitochondrial ribosomal protein S16 |
| chr13_+_66904914 | 0.34 |
ENSMUST00000168767.2
|
Gm10767
|
predicted gene 10767 |
| chr7_+_133637686 | 0.34 |
ENSMUST00000128901.2
|
2700050L05Rik
|
RIKEN cDNA 2700050L05 gene |
| chr1_-_58695944 | 0.34 |
ENSMUST00000055313.7
|
Als2cr12
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 12 (human) |
| chr14_-_30353468 | 0.33 |
ENSMUST00000112249.1
|
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr1_-_52490736 | 0.33 |
ENSMUST00000170269.1
|
Nab1
|
Ngfi-A binding protein 1 |
| chr13_-_23698454 | 0.33 |
ENSMUST00000102967.1
|
Hist1h4c
|
histone cluster 1, H4c |
| chr9_+_87022014 | 0.33 |
ENSMUST00000168529.2
ENSMUST00000174724.1 ENSMUST00000173126.1 |
Cyb5r4
|
cytochrome b5 reductase 4 |
| chr2_-_119541513 | 0.33 |
ENSMUST00000171024.1
|
Exd1
|
exonuclease 3'-5' domain containing 1 |
| chr7_+_127470354 | 0.32 |
ENSMUST00000106292.1
|
Prr14
|
proline rich 14 |
| chr7_-_130772652 | 0.32 |
ENSMUST00000057134.4
|
Etos1
|
ectopic ossification 1 |
| chr12_-_59061425 | 0.32 |
ENSMUST00000021380.8
|
Trappc6b
|
trafficking protein particle complex 6B |
| chr12_-_54695829 | 0.31 |
ENSMUST00000162106.1
ENSMUST00000160085.1 ENSMUST00000161592.1 ENSMUST00000163433.1 |
Eapp
|
E2F-associated phosphoprotein |
| chrY_+_80135210 | 0.31 |
ENSMUST00000179811.1
|
Gm21760
|
predicted gene, 21760 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Homez
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.6 | 3.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.4 | 1.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 4.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 1.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) female meiosis sister chromatid cohesion(GO:0007066) |
| 0.3 | 0.9 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.3 | 1.5 | GO:0070178 | D-serine metabolic process(GO:0070178) |
| 0.3 | 0.8 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.3 | 1.1 | GO:0042197 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.3 | 1.0 | GO:0036233 | glycine import(GO:0036233) |
| 0.2 | 0.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.8 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.2 | 0.6 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 1.1 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.2 | 0.9 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.4 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 | 1.0 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.1 | 0.4 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.1 | 0.5 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.8 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 1.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 2.9 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.3 | GO:0009804 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.8 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 0.4 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 1.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.7 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.2 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.1 | 0.4 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.4 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 0.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.4 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.5 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 0.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.7 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.3 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.7 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.4 | GO:0042559 | pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.1 | 1.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.6 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 0.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.3 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.3 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.0 | 0.8 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.5 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.3 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 1.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.8 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 1.9 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.0 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.3 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.1 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.1 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.6 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.4 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.2 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.9 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 2.5 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.8 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.1 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.0 | 0.1 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.0 | 0.6 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 1.1 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0000802 | transverse filament(GO:0000802) |
| 0.3 | 0.9 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 2.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.8 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 1.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.9 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.8 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.8 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.4 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.6 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.6 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 1.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.8 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.5 | 1.5 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.4 | 2.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 0.8 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.3 | 3.8 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 0.5 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 3.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 4.0 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 0.4 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 1.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.8 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 2.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.6 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.5 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.3 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.3 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.1 | 1.0 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.2 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 1.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.3 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.1 | 0.2 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.1 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.1 | 0.8 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.3 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 3.2 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.5 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.4 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.4 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.6 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 0.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 2.1 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.7 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |