Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
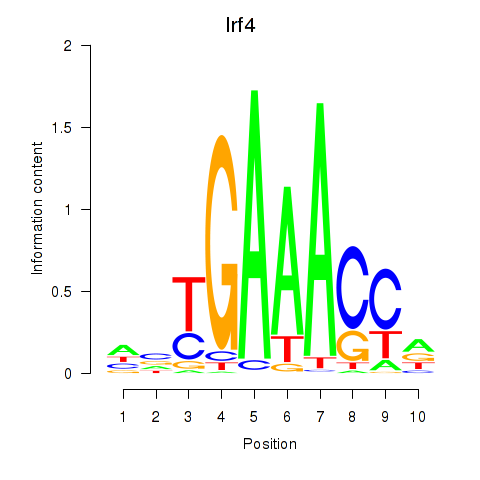
Results for Irf4
Z-value: 0.31
Transcription factors associated with Irf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf4
|
ENSMUSG00000021356.3 | interferon regulatory factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf4 | mm10_v2_chr13_+_30749226_30749247 | 0.07 | 7.0e-01 | Click! |
Activity profile of Irf4 motif
Sorted Z-values of Irf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_53618659 | 0.36 |
ENSMUST00000000889.6
|
Il4
|
interleukin 4 |
| chr19_+_34640871 | 0.30 |
ENSMUST00000102824.3
|
Ifit1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr7_-_126594941 | 0.29 |
ENSMUST00000058429.5
|
Il27
|
interleukin 27 |
| chr8_+_84723003 | 0.29 |
ENSMUST00000098571.4
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr13_+_19214103 | 0.28 |
ENSMUST00000103558.1
|
Tcrg-C1
|
T cell receptor gamma, constant 1 |
| chr5_-_66514815 | 0.27 |
ENSMUST00000161879.1
ENSMUST00000159357.1 |
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chrX_+_49463926 | 0.27 |
ENSMUST00000130558.1
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr7_+_35802593 | 0.24 |
ENSMUST00000052454.2
|
E130304I02Rik
|
RIKEN cDNA E130304I02 gene |
| chr3_+_142496924 | 0.24 |
ENSMUST00000090127.2
|
Gbp5
|
guanylate binding protein 5 |
| chr2_-_52335134 | 0.23 |
ENSMUST00000075301.3
|
Neb
|
nebulin |
| chr1_-_153851189 | 0.23 |
ENSMUST00000059607.6
|
5830403L16Rik
|
RIKEN cDNA 5830403L16 gene |
| chr11_-_95699143 | 0.21 |
ENSMUST00000062249.2
|
Gm9796
|
predicted gene 9796 |
| chr19_+_41029275 | 0.20 |
ENSMUST00000051806.4
ENSMUST00000112200.1 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr4_-_133263042 | 0.19 |
ENSMUST00000105908.3
ENSMUST00000030674.7 |
Sytl1
|
synaptotagmin-like 1 |
| chr19_+_55180799 | 0.19 |
ENSMUST00000025936.5
|
Tectb
|
tectorin beta |
| chr1_-_150164943 | 0.18 |
ENSMUST00000181308.1
|
Gm26687
|
predicted gene, 26687 |
| chrX_-_134161928 | 0.17 |
ENSMUST00000033611.4
|
Xkrx
|
X Kell blood group precursor related X linked |
| chr10_-_39163794 | 0.16 |
ENSMUST00000076713.4
|
Wisp3
|
WNT1 inducible signaling pathway protein 3 |
| chr7_+_4460687 | 0.16 |
ENSMUST00000167298.1
ENSMUST00000171445.1 |
Eps8l1
|
EPS8-like 1 |
| chr10_+_28668360 | 0.15 |
ENSMUST00000060409.6
ENSMUST00000056097.4 ENSMUST00000105516.2 |
Themis
|
thymocyte selection associated |
| chr7_-_141266415 | 0.15 |
ENSMUST00000106023.1
ENSMUST00000097952.2 ENSMUST00000026571.4 |
Irf7
|
interferon regulatory factor 7 |
| chr11_+_65162132 | 0.15 |
ENSMUST00000181156.1
|
B430202K04Rik
|
RIKEN cDNA B430202K04 gene |
| chr12_-_26456423 | 0.15 |
ENSMUST00000020970.7
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr2_-_72813665 | 0.14 |
ENSMUST00000136807.1
ENSMUST00000148327.1 |
6430710C18Rik
|
RIKEN cDNA 6430710C18 gene |
| chrX_+_150547375 | 0.14 |
ENSMUST00000066337.6
ENSMUST00000112715.1 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr9_+_64811313 | 0.14 |
ENSMUST00000038890.5
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr9_+_5345450 | 0.14 |
ENSMUST00000151332.1
|
Casp12
|
caspase 12 |
| chr6_+_40110251 | 0.14 |
ENSMUST00000061740.7
|
Tmem178b
|
transmembrane protein 178B |
| chr14_+_120275669 | 0.13 |
ENSMUST00000088419.6
ENSMUST00000167459.1 |
Mbnl2
|
muscleblind-like 2 |
| chr9_-_111057235 | 0.13 |
ENSMUST00000111888.1
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr13_-_100552704 | 0.13 |
ENSMUST00000022140.5
|
Ocln
|
occludin |
| chr13_-_100552598 | 0.12 |
ENSMUST00000159459.1
|
Ocln
|
occludin |
| chrX_-_108834303 | 0.12 |
ENSMUST00000101283.3
ENSMUST00000150434.1 |
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr19_-_57197435 | 0.12 |
ENSMUST00000111550.1
|
Ablim1
|
actin-binding LIM protein 1 |
| chr17_-_81649607 | 0.12 |
ENSMUST00000163680.2
ENSMUST00000086538.3 ENSMUST00000163123.1 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr7_+_105375053 | 0.11 |
ENSMUST00000106805.2
|
Gm5901
|
predicted gene 5901 |
| chr19_-_57197496 | 0.11 |
ENSMUST00000111544.1
|
Ablim1
|
actin-binding LIM protein 1 |
| chr13_-_100552442 | 0.11 |
ENSMUST00000159515.1
ENSMUST00000160859.1 ENSMUST00000069756.4 |
Ocln
|
occludin |
| chr13_+_24614608 | 0.11 |
ENSMUST00000091694.3
|
Fam65b
|
family with sequence similarity 65, member B |
| chr18_-_78142119 | 0.11 |
ENSMUST00000160639.1
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr19_+_55180716 | 0.11 |
ENSMUST00000154886.1
ENSMUST00000120936.1 |
Tectb
|
tectorin beta |
| chr19_-_57197377 | 0.11 |
ENSMUST00000111546.1
|
Ablim1
|
actin-binding LIM protein 1 |
| chr8_-_68121527 | 0.11 |
ENSMUST00000178529.1
|
Gm21807
|
predicted gene, 21807 |
| chr13_-_18031616 | 0.10 |
ENSMUST00000099736.2
|
Vdac3-ps1
|
voltage-dependent anion channel 3, pseudogene 1 |
| chr9_-_77251829 | 0.10 |
ENSMUST00000184322.1
ENSMUST00000184316.1 |
Mlip
|
muscular LMNA-interacting protein |
| chr11_+_82115180 | 0.10 |
ENSMUST00000009329.2
|
Ccl8
|
chemokine (C-C motif) ligand 8 |
| chr2_+_23069210 | 0.10 |
ENSMUST00000155602.1
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr3_-_30699843 | 0.10 |
ENSMUST00000108263.1
|
Lrrc31
|
leucine rich repeat containing 31 |
| chr9_-_77251871 | 0.10 |
ENSMUST00000183955.1
|
Mlip
|
muscular LMNA-interacting protein |
| chr7_-_80387935 | 0.10 |
ENSMUST00000080932.6
|
Fes
|
feline sarcoma oncogene |
| chr4_-_156200818 | 0.10 |
ENSMUST00000085425.4
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr12_-_110695860 | 0.10 |
ENSMUST00000149189.1
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr15_-_66560997 | 0.09 |
ENSMUST00000048372.5
|
Tmem71
|
transmembrane protein 71 |
| chr11_+_49087022 | 0.09 |
ENSMUST00000046704.6
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr17_+_35424870 | 0.09 |
ENSMUST00000113879.3
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr10_+_94575257 | 0.08 |
ENSMUST00000121471.1
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chrX_-_9469288 | 0.08 |
ENSMUST00000015484.3
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr1_+_132008285 | 0.08 |
ENSMUST00000146432.1
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr5_+_65863563 | 0.08 |
ENSMUST00000031106.4
|
Rhoh
|
ras homolog gene family, member H |
| chr4_+_115000174 | 0.08 |
ENSMUST00000129957.1
|
Stil
|
Scl/Tal1 interrupting locus |
| chr16_-_43664145 | 0.08 |
ENSMUST00000096065.4
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains |
| chr11_+_67321778 | 0.08 |
ENSMUST00000180845.1
|
Myh13
|
myosin, heavy polypeptide 13, skeletal muscle |
| chr11_+_67321638 | 0.08 |
ENSMUST00000108684.1
|
Myh13
|
myosin, heavy polypeptide 13, skeletal muscle |
| chrM_+_8600 | 0.08 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr19_+_34607927 | 0.08 |
ENSMUST00000076249.5
|
I830012O16Rik
|
RIKEN cDNA I830012O16 gene |
| chr19_+_34583528 | 0.08 |
ENSMUST00000102825.3
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr3_+_95526777 | 0.07 |
ENSMUST00000015667.2
ENSMUST00000116304.2 |
Ctss
|
cathepsin S |
| chrX_+_159303266 | 0.07 |
ENSMUST00000112491.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr7_+_19024387 | 0.07 |
ENSMUST00000153976.1
|
Sympk
|
symplekin |
| chr11_+_119393060 | 0.07 |
ENSMUST00000131035.2
ENSMUST00000093902.5 |
Rnf213
|
ring finger protein 213 |
| chr4_+_115000156 | 0.07 |
ENSMUST00000030490.6
|
Stil
|
Scl/Tal1 interrupting locus |
| chr2_-_173218879 | 0.07 |
ENSMUST00000109116.2
ENSMUST00000029018.7 |
Zbp1
|
Z-DNA binding protein 1 |
| chr19_-_57197556 | 0.07 |
ENSMUST00000099294.2
|
Ablim1
|
actin-binding LIM protein 1 |
| chr7_-_103840427 | 0.07 |
ENSMUST00000106866.1
|
Hbb-bh2
|
hemoglobin beta, bh2 |
| chr16_-_26371828 | 0.07 |
ENSMUST00000023154.2
|
Cldn1
|
claudin 1 |
| chr3_+_19989151 | 0.06 |
ENSMUST00000173779.1
|
Cp
|
ceruloplasmin |
| chr17_+_8849974 | 0.06 |
ENSMUST00000115720.1
|
Pde10a
|
phosphodiesterase 10A |
| chrM_+_9452 | 0.06 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr11_+_62820469 | 0.06 |
ENSMUST00000108703.1
|
Trim16
|
tripartite motif-containing 16 |
| chr1_-_159168642 | 0.06 |
ENSMUST00000077309.1
|
Gm4953
|
predicted pseudogene 4953 |
| chr7_+_35586232 | 0.06 |
ENSMUST00000040844.9
|
Ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr1_-_97977233 | 0.06 |
ENSMUST00000161567.1
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr10_+_126978690 | 0.06 |
ENSMUST00000105256.2
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr1_-_57406443 | 0.06 |
ENSMUST00000160837.1
ENSMUST00000161780.1 |
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr3_+_68691424 | 0.05 |
ENSMUST00000107816.2
|
Il12a
|
interleukin 12a |
| chr19_-_58455161 | 0.05 |
ENSMUST00000135730.1
ENSMUST00000152507.1 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr11_+_88047788 | 0.05 |
ENSMUST00000107920.3
|
Srsf1
|
serine/arginine-rich splicing factor 1 |
| chr15_-_50889691 | 0.05 |
ENSMUST00000165201.2
ENSMUST00000184458.1 |
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr13_-_64497792 | 0.05 |
ENSMUST00000180282.1
|
1190003K10Rik
|
RIKEN cDNA 1190003K10 gene |
| chr10_-_100589205 | 0.05 |
ENSMUST00000054471.8
|
4930430F08Rik
|
RIKEN cDNA 4930430F08 gene |
| chr7_+_127296369 | 0.05 |
ENSMUST00000170971.1
|
Itgal
|
integrin alpha L |
| chr7_-_65370908 | 0.05 |
ENSMUST00000032729.6
|
Tjp1
|
tight junction protein 1 |
| chr10_-_111997204 | 0.05 |
ENSMUST00000074805.5
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr7_+_78914216 | 0.05 |
ENSMUST00000120331.2
|
Isg20
|
interferon-stimulated protein |
| chr2_+_128126030 | 0.05 |
ENSMUST00000089634.5
ENSMUST00000019281.7 ENSMUST00000110341.2 ENSMUST00000103211.1 ENSMUST00000103210.1 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr2_-_131174653 | 0.05 |
ENSMUST00000127987.1
|
Spef1
|
sperm flagellar 1 |
| chr13_-_100616911 | 0.05 |
ENSMUST00000168772.1
ENSMUST00000163163.1 ENSMUST00000022137.7 |
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chr2_-_131175201 | 0.05 |
ENSMUST00000110218.2
|
Spef1
|
sperm flagellar 1 |
| chr13_+_42052015 | 0.04 |
ENSMUST00000060148.5
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr12_-_79007276 | 0.04 |
ENSMUST00000056660.6
ENSMUST00000174721.1 |
Tmem229b
|
transmembrane protein 229B |
| chr12_+_111417430 | 0.04 |
ENSMUST00000072646.6
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr3_+_3634145 | 0.04 |
ENSMUST00000108394.1
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr14_-_122465677 | 0.04 |
ENSMUST00000039118.6
|
Zic5
|
zinc finger protein of the cerebellum 5 |
| chr17_+_34564268 | 0.04 |
ENSMUST00000015612.7
|
Notch4
|
notch 4 |
| chr9_+_5345414 | 0.04 |
ENSMUST00000027009.4
|
Casp12
|
caspase 12 |
| chr10_+_86022189 | 0.04 |
ENSMUST00000120344.1
ENSMUST00000117597.1 |
Fbxo7
|
F-box protein 7 |
| chr1_-_74588117 | 0.04 |
ENSMUST00000066986.6
|
Zfp142
|
zinc finger protein 142 |
| chr6_+_35177386 | 0.04 |
ENSMUST00000043815.9
|
Nup205
|
nucleoporin 205 |
| chrX_+_112615301 | 0.04 |
ENSMUST00000122805.1
|
Zfp711
|
zinc finger protein 711 |
| chr2_-_152830615 | 0.04 |
ENSMUST00000146380.1
ENSMUST00000134902.1 ENSMUST00000134357.1 ENSMUST00000109820.3 |
Bcl2l1
|
BCL2-like 1 |
| chr18_-_70530138 | 0.04 |
ENSMUST00000161542.1
ENSMUST00000159389.1 |
Poli
|
polymerase (DNA directed), iota |
| chr10_-_64090241 | 0.04 |
ENSMUST00000133588.1
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr16_-_55838827 | 0.04 |
ENSMUST00000096026.2
ENSMUST00000036273.6 ENSMUST00000114457.1 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr10_+_69212676 | 0.04 |
ENSMUST00000167384.1
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr7_+_19024364 | 0.04 |
ENSMUST00000023882.7
|
Sympk
|
symplekin |
| chr7_+_127296291 | 0.04 |
ENSMUST00000106306.2
ENSMUST00000120857.1 |
Itgal
|
integrin alpha L |
| chr2_-_152831112 | 0.04 |
ENSMUST00000128172.1
|
Bcl2l1
|
BCL2-like 1 |
| chr7_+_127296251 | 0.04 |
ENSMUST00000117762.1
|
Itgal
|
integrin alpha L |
| chr13_+_23555023 | 0.04 |
ENSMUST00000045301.6
|
Hist1h1d
|
histone cluster 1, H1d |
| chr11_-_120348513 | 0.03 |
ENSMUST00000071555.6
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr6_+_71282280 | 0.03 |
ENSMUST00000080949.7
|
Krcc1
|
lysine-rich coiled-coil 1 |
| chr6_-_124911636 | 0.03 |
ENSMUST00000032217.1
|
Lag3
|
lymphocyte-activation gene 3 |
| chr1_-_161876656 | 0.03 |
ENSMUST00000048377.5
|
Suco
|
SUN domain containing ossification factor |
| chr1_+_57406328 | 0.03 |
ENSMUST00000027114.5
|
9430016H08Rik
|
RIKEN cDNA 9430016H08 gene |
| chr1_+_66468364 | 0.03 |
ENSMUST00000061620.9
|
Unc80
|
unc-80 homolog (C. elegans) |
| chr3_+_3508024 | 0.03 |
ENSMUST00000108393.1
ENSMUST00000017832.8 |
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr1_-_191164815 | 0.03 |
ENSMUST00000171798.1
|
Fam71a
|
family with sequence similarity 71, member A |
| chr7_+_58658181 | 0.03 |
ENSMUST00000168747.1
|
Atp10a
|
ATPase, class V, type 10A |
| chr2_+_23069057 | 0.03 |
ENSMUST00000114526.1
ENSMUST00000114529.2 |
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr11_-_43836243 | 0.03 |
ENSMUST00000167574.1
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chr15_-_76243401 | 0.03 |
ENSMUST00000165738.1
ENSMUST00000075689.6 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chrX_+_85870324 | 0.03 |
ENSMUST00000113976.1
|
5430427O19Rik
|
RIKEN cDNA 5430427O19 gene |
| chr18_-_70530313 | 0.03 |
ENSMUST00000043286.8
|
Poli
|
polymerase (DNA directed), iota |
| chr9_+_98864767 | 0.03 |
ENSMUST00000167951.1
|
2410012M07Rik
|
RIKEN cDNA 2410012M07 gene |
| chr9_-_117251801 | 0.03 |
ENSMUST00000172564.1
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr1_+_187215737 | 0.03 |
ENSMUST00000160471.1
|
Gpatch2
|
G patch domain containing 2 |
| chr13_+_20090538 | 0.02 |
ENSMUST00000072519.5
|
Elmo1
|
engulfment and cell motility 1 |
| chr18_+_7869707 | 0.02 |
ENSMUST00000166062.1
ENSMUST00000169010.1 |
Wac
|
WW domain containing adaptor with coiled-coil |
| chr11_+_46810792 | 0.02 |
ENSMUST00000068877.6
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr1_-_75316998 | 0.02 |
ENSMUST00000113605.3
|
Dnpep
|
aspartyl aminopeptidase |
| chr14_+_55578360 | 0.02 |
ENSMUST00000174259.1
ENSMUST00000174563.1 ENSMUST00000089619.6 ENSMUST00000172738.1 |
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr8_+_90828820 | 0.02 |
ENSMUST00000109614.2
ENSMUST00000048665.6 |
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr1_-_93101825 | 0.02 |
ENSMUST00000112958.2
|
Kif1a
|
kinesin family member 1A |
| chr8_+_61928081 | 0.02 |
ENSMUST00000154398.1
ENSMUST00000093485.2 ENSMUST00000156980.1 ENSMUST00000070631.7 |
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr13_+_20090500 | 0.02 |
ENSMUST00000165249.2
|
Elmo1
|
engulfment and cell motility 1 |
| chr10_+_128706251 | 0.02 |
ENSMUST00000054125.8
|
Pmel
|
premelanosome protein |
| chr9_-_116175318 | 0.02 |
ENSMUST00000061101.4
ENSMUST00000035014.6 |
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr1_+_155558112 | 0.02 |
ENSMUST00000080138.6
ENSMUST00000097529.3 ENSMUST00000035560.3 |
Acbd6
|
acyl-Coenzyme A binding domain containing 6 |
| chr10_-_18234930 | 0.02 |
ENSMUST00000052648.8
ENSMUST00000080860.6 ENSMUST00000173243.1 |
Ccdc28a
|
coiled-coil domain containing 28A |
| chr19_+_46623387 | 0.01 |
ENSMUST00000111855.4
|
Wbp1l
|
WW domain binding protein 1 like |
| chr4_-_46536096 | 0.01 |
ENSMUST00000102924.2
|
Trim14
|
tripartite motif-containing 14 |
| chr1_+_188263034 | 0.01 |
ENSMUST00000060479.7
|
Ush2a
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr11_+_87566653 | 0.01 |
ENSMUST00000060360.5
|
Gm11492
|
predicted gene 11492 |
| chr2_+_29935413 | 0.01 |
ENSMUST00000019859.8
|
Gle1
|
GLE1 RNA export mediator (yeast) |
| chr1_-_93101854 | 0.01 |
ENSMUST00000171796.1
ENSMUST00000171556.1 |
Kif1a
|
kinesin family member 1A |
| chr10_-_107719978 | 0.01 |
ENSMUST00000050702.7
|
Ptprq
|
protein tyrosine phosphatase, receptor type, Q |
| chr6_-_39118211 | 0.01 |
ENSMUST00000038398.6
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr11_-_69920581 | 0.01 |
ENSMUST00000108610.1
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr1_+_82724881 | 0.01 |
ENSMUST00000078332.6
|
Mff
|
mitochondrial fission factor |
| chr10_+_69212634 | 0.01 |
ENSMUST00000020101.5
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr17_+_47410247 | 0.01 |
ENSMUST00000177586.1
|
Gm5814
|
predicted pseudogene 5814 |
| chr4_-_72200833 | 0.01 |
ENSMUST00000102848.2
ENSMUST00000072695.6 ENSMUST00000107337.1 ENSMUST00000074216.7 |
Tle1
|
transducin-like enhancer of split 1, homolog of Drosophila E(spl) |
| chr10_-_86022325 | 0.01 |
ENSMUST00000181665.1
|
A230060F14Rik
|
RIKEN cDNA A230060F14 gene |
| chrX_+_94367112 | 0.01 |
ENSMUST00000113898.1
|
Apoo
|
apolipoprotein O |
| chr14_+_55578123 | 0.01 |
ENSMUST00000174484.1
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr13_+_5861489 | 0.01 |
ENSMUST00000000080.6
|
Klf6
|
Kruppel-like factor 6 |
| chr16_+_65520503 | 0.01 |
ENSMUST00000176330.1
ENSMUST00000004964.8 ENSMUST00000176038.1 |
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr3_-_108200807 | 0.01 |
ENSMUST00000106655.1
ENSMUST00000065664.6 |
Cyb561d1
|
cytochrome b-561 domain containing 1 |
| chr7_+_83755904 | 0.01 |
ENSMUST00000051522.8
ENSMUST00000042280.7 |
Gm7964
|
predicted gene 7964 |
| chr16_-_5255923 | 0.01 |
ENSMUST00000139584.1
ENSMUST00000064635.5 |
Fam86
|
family with sequence similarity 86 |
| chr17_-_6782775 | 0.01 |
ENSMUST00000064234.6
|
Ezr
|
ezrin |
| chr18_+_37484955 | 0.01 |
ENSMUST00000053856.4
|
Pcdhb17
|
protocadherin beta 17 |
| chr9_+_108508005 | 0.01 |
ENSMUST00000006838.8
ENSMUST00000134939.1 |
Qars
|
glutaminyl-tRNA synthetase |
| chr9_+_35267857 | 0.01 |
ENSMUST00000034543.4
|
Rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr19_-_24225015 | 0.01 |
ENSMUST00000099558.4
|
Tjp2
|
tight junction protein 2 |
| chr4_-_107178282 | 0.01 |
ENSMUST00000058585.7
|
Tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr9_+_105053239 | 0.01 |
ENSMUST00000035177.8
ENSMUST00000149243.1 |
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chr14_-_66124482 | 0.00 |
ENSMUST00000070515.1
|
Ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr7_-_16286744 | 0.00 |
ENSMUST00000150528.2
ENSMUST00000118976.2 ENSMUST00000146609.2 |
Ccdc9
|
coiled-coil domain containing 9 |
| chrX_+_20848543 | 0.00 |
ENSMUST00000001155.4
ENSMUST00000122312.1 ENSMUST00000120356.1 ENSMUST00000122850.1 |
Araf
|
v-raf murine sarcoma 3611 viral oncogene homolog |
| chr6_+_128362919 | 0.00 |
ENSMUST00000073316.6
|
Foxm1
|
forkhead box M1 |
| chr11_+_53720790 | 0.00 |
ENSMUST00000048605.2
|
Il5
|
interleukin 5 |
| chr19_+_47937648 | 0.00 |
ENSMUST00000066308.7
|
Ccdc147
|
coiled-coil domain containing 147 |
| chr8_-_125492710 | 0.00 |
ENSMUST00000108775.1
|
Sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr4_-_129239165 | 0.00 |
ENSMUST00000097873.3
|
C77080
|
expressed sequence C77080 |
| chr2_+_180589245 | 0.00 |
ENSMUST00000029087.3
|
Ogfr
|
opioid growth factor receptor |
| chr17_-_30612613 | 0.00 |
ENSMUST00000167624.1
ENSMUST00000024823.6 |
Glo1
|
glyoxalase 1 |
| chr15_+_52295509 | 0.00 |
ENSMUST00000037240.2
|
Slc30a8
|
solute carrier family 30 (zinc transporter), member 8 |
| chrX_+_94636066 | 0.00 |
ENSMUST00000096368.3
|
Gspt2
|
G1 to S phase transition 2 |
| chr9_+_65346066 | 0.00 |
ENSMUST00000048184.2
|
Pdcd7
|
programmed cell death 7 |
| chr1_-_156939387 | 0.00 |
ENSMUST00000171292.1
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr4_-_46536134 | 0.00 |
ENSMUST00000046897.6
|
Trim14
|
tripartite motif-containing 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.2 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.2 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.4 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.1 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.1 | GO:0098735 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.0 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.0 | 0.1 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.1 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.0 | 0.1 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |