Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
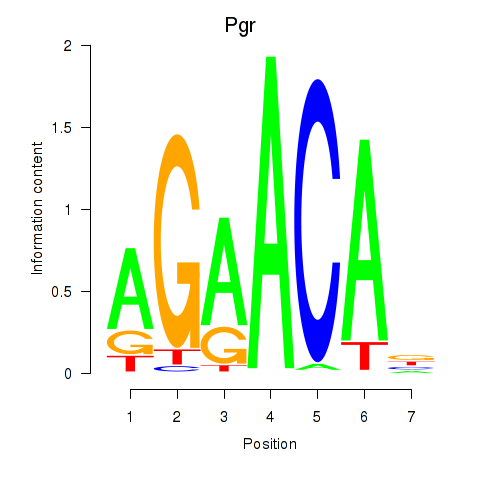
Results for Pgr_Nr3c1
Z-value: 1.40
Transcription factors associated with Pgr_Nr3c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pgr
|
ENSMUSG00000031870.10 | progesterone receptor |
|
Nr3c1
|
ENSMUSG00000024431.8 | nuclear receptor subfamily 3, group C, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pgr | mm10_v2_chr9_+_8900459_8900489 | -0.47 | 4.1e-03 | Click! |
| Nr3c1 | mm10_v2_chr18_-_39489880_39489915 | -0.24 | 1.5e-01 | Click! |
Activity profile of Pgr_Nr3c1 motif
Sorted Z-values of Pgr_Nr3c1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_30924169 | 13.89 |
ENSMUST00000074671.6
|
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr6_+_125320633 | 8.41 |
ENSMUST00000176655.1
ENSMUST00000176110.1 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr6_-_138079916 | 4.78 |
ENSMUST00000171804.1
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr16_-_10543028 | 4.75 |
ENSMUST00000184863.1
ENSMUST00000038281.5 |
Dexi
|
dexamethasone-induced transcript |
| chr16_+_43363855 | 3.51 |
ENSMUST00000156367.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chrX_+_101383726 | 3.28 |
ENSMUST00000119190.1
|
Gjb1
|
gap junction protein, beta 1 |
| chr10_-_109010955 | 2.91 |
ENSMUST00000105276.1
ENSMUST00000064054.7 |
Syt1
|
synaptotagmin I |
| chr5_-_103629279 | 2.88 |
ENSMUST00000031263.1
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr6_-_124542281 | 2.82 |
ENSMUST00000159463.1
ENSMUST00000162844.1 ENSMUST00000160505.1 ENSMUST00000162443.1 |
C1s
|
complement component 1, s subcomponent |
| chr11_+_117825933 | 2.73 |
ENSMUST00000149668.1
|
Afmid
|
arylformamidase |
| chr11_+_117825885 | 2.64 |
ENSMUST00000073388.6
|
Afmid
|
arylformamidase |
| chr6_-_35326123 | 2.54 |
ENSMUST00000051176.7
|
Fam180a
|
family with sequence similarity 180, member A |
| chr13_+_24943144 | 2.43 |
ENSMUST00000021773.5
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr5_-_66080971 | 2.39 |
ENSMUST00000127275.1
ENSMUST00000113724.1 |
Rbm47
|
RNA binding motif protein 47 |
| chr17_-_34743849 | 2.33 |
ENSMUST00000069507.8
|
C4b
|
complement component 4B (Chido blood group) |
| chr6_+_125321205 | 2.30 |
ENSMUST00000176365.1
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr18_+_9707639 | 2.06 |
ENSMUST00000040069.8
|
Colec12
|
collectin sub-family member 12 |
| chr6_+_125321333 | 1.94 |
ENSMUST00000081440.7
|
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr1_-_171240055 | 1.92 |
ENSMUST00000131286.1
|
Ndufs2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2 |
| chr5_+_30814571 | 1.90 |
ENSMUST00000031058.8
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr5_+_30814722 | 1.82 |
ENSMUST00000114724.1
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chrX_+_21714896 | 1.81 |
ENSMUST00000033414.7
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr1_+_93235836 | 1.79 |
ENSMUST00000062202.7
|
Sned1
|
sushi, nidogen and EGF-like domains 1 |
| chr5_+_65131184 | 1.74 |
ENSMUST00000031089.5
ENSMUST00000101191.3 |
Klhl5
|
kelch-like 5 |
| chr14_+_123659971 | 1.71 |
ENSMUST00000049681.7
|
Itgbl1
|
integrin, beta-like 1 |
| chr3_-_95904683 | 1.70 |
ENSMUST00000147962.1
ENSMUST00000036181.8 |
Car14
|
carbonic anhydrase 14 |
| chr19_-_42202150 | 1.66 |
ENSMUST00000018966.7
|
Sfrp5
|
secreted frizzled-related sequence protein 5 |
| chr17_+_8165501 | 1.51 |
ENSMUST00000097419.3
ENSMUST00000024636.8 |
Fgfr1op
|
Fgfr1 oncogene partner |
| chr2_-_77170534 | 1.45 |
ENSMUST00000111833.2
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr2_-_103303158 | 1.44 |
ENSMUST00000111176.2
|
Ehf
|
ets homologous factor |
| chr11_+_4883186 | 1.43 |
ENSMUST00000139737.1
|
Nipsnap1
|
4-nitrophenylphosphatase domain and non-neuronal SNAP25-like protein homolog 1 (C. elegans) |
| chr4_+_99295900 | 1.42 |
ENSMUST00000094955.1
|
Gm12689
|
predicted gene 12689 |
| chr15_-_50890396 | 1.37 |
ENSMUST00000185183.1
|
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr3_-_57692537 | 1.37 |
ENSMUST00000099091.3
|
Gm410
|
predicted gene 410 |
| chr8_-_3717547 | 1.33 |
ENSMUST00000058040.6
|
Gm9814
|
predicted gene 9814 |
| chr18_+_20665250 | 1.32 |
ENSMUST00000075312.3
|
Ttr
|
transthyretin |
| chr17_-_34214459 | 1.30 |
ENSMUST00000121995.1
|
Gm15821
|
predicted gene 15821 |
| chr14_+_70555900 | 1.27 |
ENSMUST00000163060.1
|
Hr
|
hairless |
| chr12_-_86892540 | 1.26 |
ENSMUST00000181290.1
|
Gm26698
|
predicted gene, 26698 |
| chr11_+_112782182 | 1.26 |
ENSMUST00000000579.2
|
Sox9
|
SRY-box containing gene 9 |
| chr5_-_24601961 | 1.23 |
ENSMUST00000030791.7
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr2_-_103303179 | 1.21 |
ENSMUST00000090475.3
|
Ehf
|
ets homologous factor |
| chr4_-_119422355 | 1.21 |
ENSMUST00000106316.1
ENSMUST00000030385.6 |
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr2_-_76647992 | 1.16 |
ENSMUST00000002808.6
|
Prkra
|
protein kinase, interferon inducible double stranded RNA dependent activator |
| chr3_-_89387132 | 1.13 |
ENSMUST00000107433.1
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr7_-_127824469 | 1.09 |
ENSMUST00000106267.3
|
Stx1b
|
syntaxin 1B |
| chr14_-_124677089 | 1.08 |
ENSMUST00000095529.3
|
Fgf14
|
fibroblast growth factor 14 |
| chr3_-_97868242 | 1.07 |
ENSMUST00000107038.3
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin) |
| chr15_-_89477400 | 1.05 |
ENSMUST00000165199.1
|
Arsa
|
arylsulfatase A |
| chr7_+_30553263 | 1.03 |
ENSMUST00000044048.7
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6 |
| chr6_-_59024470 | 1.02 |
ENSMUST00000089860.5
|
Fam13a
|
family with sequence similarity 13, member A |
| chr6_+_37870786 | 0.93 |
ENSMUST00000120428.1
ENSMUST00000031859.7 |
Trim24
|
tripartite motif-containing 24 |
| chr19_-_6084679 | 0.92 |
ENSMUST00000161548.1
|
Zfpl1
|
zinc finger like protein 1 |
| chr5_+_125441546 | 0.90 |
ENSMUST00000049040.9
|
Bri3bp
|
Bri3 binding protein |
| chr2_-_63184253 | 0.90 |
ENSMUST00000075052.3
ENSMUST00000112454.1 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr15_-_65912254 | 0.89 |
ENSMUST00000079776.7
ENSMUST00000060522.4 |
Oc90
|
otoconin 90 |
| chr2_+_91255954 | 0.88 |
ENSMUST00000134699.1
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr2_-_63184170 | 0.88 |
ENSMUST00000112452.1
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr2_+_91256144 | 0.88 |
ENSMUST00000154959.1
ENSMUST00000059566.4 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr6_-_59024340 | 0.85 |
ENSMUST00000173193.1
|
Fam13a
|
family with sequence similarity 13, member A |
| chr15_-_81399594 | 0.83 |
ENSMUST00000023039.8
|
St13
|
suppression of tumorigenicity 13 |
| chr11_-_3931789 | 0.83 |
ENSMUST00000109992.1
ENSMUST00000109988.1 |
Tcn2
|
transcobalamin 2 |
| chr14_-_69732510 | 0.81 |
ENSMUST00000036381.8
|
Chmp7
|
charged multivesicular body protein 7 |
| chr11_+_83709015 | 0.78 |
ENSMUST00000001009.7
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr15_-_98898483 | 0.77 |
ENSMUST00000023737.4
|
Dhh
|
desert hedgehog |
| chr3_-_33082004 | 0.75 |
ENSMUST00000108225.3
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr3_+_121531603 | 0.71 |
ENSMUST00000180804.1
|
A530020G20Rik
|
RIKEN cDNA A530020G20 gene |
| chr19_-_55315980 | 0.70 |
ENSMUST00000076891.5
|
Zdhhc6
|
zinc finger, DHHC domain containing 6 |
| chr11_-_3931960 | 0.70 |
ENSMUST00000109990.1
ENSMUST00000020710.4 ENSMUST00000109989.3 ENSMUST00000109991.1 ENSMUST00000109993.2 |
Tcn2
|
transcobalamin 2 |
| chr4_-_141618238 | 0.69 |
ENSMUST00000053263.8
|
Tmem82
|
transmembrane protein 82 |
| chr19_-_6084873 | 0.68 |
ENSMUST00000160977.1
ENSMUST00000159859.1 |
Zfpl1
|
zinc finger like protein 1 |
| chr11_-_121519326 | 0.66 |
ENSMUST00000092298.5
|
Zfp750
|
zinc finger protein 750 |
| chr3_-_97868191 | 0.64 |
ENSMUST00000163531.2
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin) |
| chr9_+_46273064 | 0.64 |
ENSMUST00000156440.1
ENSMUST00000034583.6 ENSMUST00000114552.3 |
Zfp259
|
zinc finger protein 259 |
| chr7_-_127345314 | 0.62 |
ENSMUST00000060783.5
|
Zfp768
|
zinc finger protein 768 |
| chr12_-_4477138 | 0.62 |
ENSMUST00000085814.3
|
Ncoa1
|
nuclear receptor coactivator 1 |
| chr16_+_21891969 | 0.61 |
ENSMUST00000042065.6
|
Map3k13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr15_-_81400043 | 0.61 |
ENSMUST00000172107.1
ENSMUST00000169204.1 ENSMUST00000163382.1 |
St13
|
suppression of tumorigenicity 13 |
| chr7_+_30712209 | 0.60 |
ENSMUST00000005692.6
ENSMUST00000170371.1 |
Atp4a
|
ATPase, H+/K+ exchanging, gastric, alpha polypeptide |
| chr10_-_107494719 | 0.58 |
ENSMUST00000044210.3
|
Myf6
|
myogenic factor 6 |
| chr10_-_116473418 | 0.57 |
ENSMUST00000087965.4
ENSMUST00000164271.1 |
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr17_-_25081138 | 0.57 |
ENSMUST00000024984.6
|
Tmem204
|
transmembrane protein 204 |
| chr11_-_70755151 | 0.56 |
ENSMUST00000127333.1
|
Gm12320
|
predicted gene 12320 |
| chr2_-_77170592 | 0.54 |
ENSMUST00000164114.2
ENSMUST00000049544.7 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr7_+_105640448 | 0.54 |
ENSMUST00000058333.3
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr8_+_123373778 | 0.54 |
ENSMUST00000057934.3
ENSMUST00000108840.2 |
Tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr12_+_103532435 | 0.53 |
ENSMUST00000021631.5
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr19_-_5797410 | 0.53 |
ENSMUST00000173314.1
|
Malat1
|
metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA) |
| chr4_-_91376433 | 0.53 |
ENSMUST00000107109.2
ENSMUST00000107111.2 ENSMUST00000107120.1 |
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) |
| chr16_-_5222257 | 0.51 |
ENSMUST00000050160.4
|
AU021092
|
expressed sequence AU021092 |
| chr11_-_66525795 | 0.51 |
ENSMUST00000123454.1
|
Shisa6
|
shisa homolog 6 (Xenopus laevis) |
| chr4_-_91376490 | 0.50 |
ENSMUST00000107124.3
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) |
| chr2_-_65567505 | 0.50 |
ENSMUST00000100069.2
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr14_+_75845093 | 0.50 |
ENSMUST00000110894.2
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr8_+_46739745 | 0.50 |
ENSMUST00000034041.7
|
Irf2
|
interferon regulatory factor 2 |
| chr6_-_72361396 | 0.50 |
ENSMUST00000130064.1
|
Rnf181
|
ring finger protein 181 |
| chr1_+_109993982 | 0.48 |
ENSMUST00000027542.6
|
Cdh7
|
cadherin 7, type 2 |
| chr3_+_87796938 | 0.47 |
ENSMUST00000029711.2
ENSMUST00000107582.2 |
Insrr
|
insulin receptor-related receptor |
| chr2_-_30474199 | 0.46 |
ENSMUST00000065134.2
|
Ier5l
|
immediate early response 5-like |
| chr11_+_88294043 | 0.46 |
ENSMUST00000037268.4
|
1700106J16Rik
|
RIKEN cDNA 1700106J16 gene |
| chr7_+_6371364 | 0.45 |
ENSMUST00000086323.4
ENSMUST00000108559.2 |
Zfp78
|
zinc finger protein 78 |
| chr4_+_88841816 | 0.44 |
ENSMUST00000094973.3
|
Ifna4
|
interferon alpha 4 |
| chr12_-_86988676 | 0.43 |
ENSMUST00000095521.2
|
Zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr2_-_167043494 | 0.43 |
ENSMUST00000067584.6
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr10_-_63421739 | 0.43 |
ENSMUST00000054760.4
|
Gm7075
|
predicted gene 7075 |
| chr19_-_45235811 | 0.42 |
ENSMUST00000099401.4
|
Lbx1
|
ladybird homeobox homolog 1 (Drosophila) |
| chr6_-_33060256 | 0.42 |
ENSMUST00000066379.4
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr6_+_17491216 | 0.42 |
ENSMUST00000080469.5
|
Met
|
met proto-oncogene |
| chr17_-_46031813 | 0.42 |
ENSMUST00000024747.7
|
Vegfa
|
vascular endothelial growth factor A |
| chr13_+_63015167 | 0.41 |
ENSMUST00000021911.8
|
2010111I01Rik
|
RIKEN cDNA 2010111I01 gene |
| chr13_-_84064772 | 0.41 |
ENSMUST00000182477.1
|
Gm17750
|
predicted gene, 17750 |
| chr7_-_63938862 | 0.41 |
ENSMUST00000063694.8
|
Klf13
|
Kruppel-like factor 13 |
| chr9_-_79718720 | 0.39 |
ENSMUST00000121227.1
|
Col12a1
|
collagen, type XII, alpha 1 |
| chr14_-_78308031 | 0.39 |
ENSMUST00000022592.7
|
Tnfsf11
|
tumor necrosis factor (ligand) superfamily, member 11 |
| chr3_-_33143025 | 0.38 |
ENSMUST00000108226.1
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr6_-_33060172 | 0.38 |
ENSMUST00000115091.1
ENSMUST00000127666.1 |
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr14_+_62760496 | 0.37 |
ENSMUST00000181344.1
|
4931440J10Rik
|
RIKEN cDNA 4931440J10 gene |
| chr6_+_32050246 | 0.37 |
ENSMUST00000031778.4
|
1700012A03Rik
|
RIKEN cDNA 1700012A03 gene |
| chr8_-_67974567 | 0.37 |
ENSMUST00000098696.3
ENSMUST00000038959.9 ENSMUST00000093469.4 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr11_+_87566653 | 0.36 |
ENSMUST00000060360.5
|
Gm11492
|
predicted gene 11492 |
| chr2_+_39008076 | 0.36 |
ENSMUST00000112862.1
ENSMUST00000090993.5 |
Arpc5l
|
actin related protein 2/3 complex, subunit 5-like |
| chr16_-_92321425 | 0.36 |
ENSMUST00000062638.5
|
4930563D23Rik
|
RIKEN cDNA 4930563D23 gene |
| chr5_-_31138829 | 0.36 |
ENSMUST00000043475.2
|
Ucn
|
urocortin |
| chr1_-_59003443 | 0.35 |
ENSMUST00000054653.6
|
Als2cr11
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11 (human) |
| chr19_+_8741669 | 0.35 |
ENSMUST00000176314.1
ENSMUST00000073430.7 ENSMUST00000175901.1 |
Stx5a
|
syntaxin 5A |
| chr4_-_141398574 | 0.34 |
ENSMUST00000133676.1
ENSMUST00000042617.7 |
Clcnka
|
chloride channel Ka |
| chr6_+_34709442 | 0.33 |
ENSMUST00000115021.1
|
Cald1
|
caldesmon 1 |
| chr4_-_110292719 | 0.32 |
ENSMUST00000106601.1
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) |
| chr1_-_9748376 | 0.31 |
ENSMUST00000057438.6
|
Vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr11_+_117332335 | 0.31 |
ENSMUST00000106349.1
|
Sept9
|
septin 9 |
| chr2_+_65620829 | 0.30 |
ENSMUST00000028377.7
|
Scn2a1
|
sodium channel, voltage-gated, type II, alpha 1 |
| chr10_-_83648631 | 0.30 |
ENSMUST00000146876.2
ENSMUST00000176294.1 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr2_+_74721978 | 0.30 |
ENSMUST00000047904.3
|
Hoxd3
|
homeobox D3 |
| chr14_-_16575456 | 0.29 |
ENSMUST00000063750.6
|
Rarb
|
retinoic acid receptor, beta |
| chr13_+_29016267 | 0.29 |
ENSMUST00000140415.1
|
A330102I10Rik
|
RIKEN cDNA A330102I10 gene |
| chr7_-_127725616 | 0.28 |
ENSMUST00000076091.2
|
Ctf2
|
cardiotrophin 2 |
| chr13_-_98637354 | 0.28 |
ENSMUST00000050389.4
|
Tmem174
|
transmembrane protein 174 |
| chr19_+_55316313 | 0.27 |
ENSMUST00000095950.2
|
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr14_+_62998063 | 0.26 |
ENSMUST00000100492.3
|
Defb47
|
defensin beta 47 |
| chr1_-_139377094 | 0.26 |
ENSMUST00000131586.1
ENSMUST00000145244.1 |
Crb1
|
crumbs homolog 1 (Drosophila) |
| chr9_+_104002546 | 0.26 |
ENSMUST00000035167.8
ENSMUST00000117054.1 |
Nphp3
|
nephronophthisis 3 (adolescent) |
| chr11_-_48816936 | 0.26 |
ENSMUST00000140800.1
|
Trim41
|
tripartite motif-containing 41 |
| chr2_-_91255995 | 0.25 |
ENSMUST00000180732.1
|
Gm17281
|
predicted gene, 17281 |
| chr8_+_124576105 | 0.25 |
ENSMUST00000093033.5
ENSMUST00000133086.1 |
Capn9
|
calpain 9 |
| chr7_+_16071942 | 0.25 |
ENSMUST00000108509.1
|
Zfp541
|
zinc finger protein 541 |
| chrX_-_50942710 | 0.23 |
ENSMUST00000060650.5
|
Frmd7
|
FERM domain containing 7 |
| chr11_+_83473079 | 0.23 |
ENSMUST00000021018.4
|
Taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr9_+_49102720 | 0.23 |
ENSMUST00000070390.5
ENSMUST00000167095.1 |
Tmprss5
|
transmembrane protease, serine 5 (spinesin) |
| chr12_-_31713873 | 0.23 |
ENSMUST00000057783.4
ENSMUST00000174480.2 ENSMUST00000176710.1 |
Gpr22
|
G protein-coupled receptor 22 |
| chr11_-_48817332 | 0.23 |
ENSMUST00000047145.7
|
Trim41
|
tripartite motif-containing 41 |
| chr14_-_63037833 | 0.23 |
ENSMUST00000111207.1
ENSMUST00000100490.2 |
Defb30
|
defensin beta 30 |
| chr6_+_106118924 | 0.22 |
ENSMUST00000079416.5
|
Cntn4
|
contactin 4 |
| chr11_+_98386450 | 0.22 |
ENSMUST00000041301.7
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr4_-_120815703 | 0.22 |
ENSMUST00000120779.1
|
Nfyc
|
nuclear transcription factor-Y gamma |
| chr2_+_163994960 | 0.22 |
ENSMUST00000018470.3
|
Ywhab
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide |
| chr1_+_71653339 | 0.22 |
ENSMUST00000125934.1
|
Apol7d
|
apolipoprotein L 7d |
| chr2_-_93996354 | 0.22 |
ENSMUST00000183110.1
|
Gm27027
|
predicted gene, 27027 |
| chr4_-_143299498 | 0.21 |
ENSMUST00000030317.7
|
Pdpn
|
podoplanin |
| chr14_+_53640946 | 0.21 |
ENSMUST00000184874.1
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chr7_-_103741322 | 0.21 |
ENSMUST00000051346.2
|
Olfr629
|
olfactory receptor 629 |
| chr8_-_46739453 | 0.21 |
ENSMUST00000181167.1
|
Gm16675
|
predicted gene, 16675 |
| chr3_-_133544390 | 0.21 |
ENSMUST00000098603.3
|
Tet2
|
tet methylcytosine dioxygenase 2 |
| chr1_-_170912941 | 0.20 |
ENSMUST00000094337.1
|
Fcrlb
|
Fc receptor-like B |
| chr6_+_21949571 | 0.20 |
ENSMUST00000031680.3
ENSMUST00000115389.1 ENSMUST00000151473.1 |
Ing3
|
inhibitor of growth family, member 3 |
| chr11_+_87664274 | 0.20 |
ENSMUST00000092800.5
|
Rnf43
|
ring finger protein 43 |
| chr14_+_52778447 | 0.20 |
ENSMUST00000103580.2
|
Trav8d-1
|
T cell receptor alpha variable 8D-1 |
| chr9_+_3404058 | 0.19 |
ENSMUST00000027027.5
|
Cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr14_+_53469756 | 0.18 |
ENSMUST00000103643.2
|
Trav8-1
|
T cell receptor alpha variable 8-1 |
| chr10_+_52388963 | 0.17 |
ENSMUST00000160539.1
|
Nepn
|
nephrocan |
| chr2_+_72297895 | 0.17 |
ENSMUST00000144111.1
|
Zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr14_+_53640701 | 0.17 |
ENSMUST00000179267.2
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chr16_-_59027927 | 0.17 |
ENSMUST00000062380.2
|
Olfr186
|
olfactory receptor 186 |
| chr15_+_81400132 | 0.16 |
ENSMUST00000163754.1
ENSMUST00000041609.4 |
Xpnpep3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr11_-_78497734 | 0.16 |
ENSMUST00000061174.6
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr12_-_73047179 | 0.15 |
ENSMUST00000050029.7
|
Six1
|
sine oculis-related homeobox 1 |
| chr4_-_136898803 | 0.13 |
ENSMUST00000046285.5
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr1_+_167001417 | 0.13 |
ENSMUST00000165874.1
|
Fam78b
|
family with sequence similarity 78, member B |
| chr7_-_30194150 | 0.13 |
ENSMUST00000126116.1
|
Capns1
|
calpain, small subunit 1 |
| chr11_-_23895208 | 0.12 |
ENSMUST00000102863.2
ENSMUST00000020513.3 |
Papolg
|
poly(A) polymerase gamma |
| chr17_-_46440099 | 0.11 |
ENSMUST00000166852.1
|
Gm5093
|
predicted gene 5093 |
| chr11_-_120630126 | 0.11 |
ENSMUST00000106180.1
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr10_+_102158858 | 0.11 |
ENSMUST00000138522.1
ENSMUST00000163753.1 ENSMUST00000138016.1 |
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr11_+_101425068 | 0.11 |
ENSMUST00000040561.5
|
Rundc1
|
RUN domain containing 1 |
| chr5_-_100373484 | 0.10 |
ENSMUST00000182433.1
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr11_-_75178792 | 0.10 |
ENSMUST00000071562.2
|
Ovca2
|
candidate tumor suppressor in ovarian cancer 2 |
| chr3_-_138067388 | 0.09 |
ENSMUST00000053318.3
|
Gm5105
|
predicted gene 5105 |
| chr17_+_34644805 | 0.09 |
ENSMUST00000174796.1
|
Fkbpl
|
FK506 binding protein-like |
| chr11_+_51027004 | 0.08 |
ENSMUST00000072152.1
|
Olfr54
|
olfactory receptor 54 |
| chr5_-_43821639 | 0.07 |
ENSMUST00000114047.3
|
Fbxl5
|
F-box and leucine-rich repeat protein 5 |
| chr18_+_77332394 | 0.07 |
ENSMUST00000148341.1
|
Loxhd1
|
lipoxygenase homology domains 1 |
| chr18_+_23415400 | 0.07 |
ENSMUST00000115832.2
ENSMUST00000047954.7 |
Dtna
|
dystrobrevin alpha |
| chr10_+_70097102 | 0.07 |
ENSMUST00000147545.1
|
Ccdc6
|
coiled-coil domain containing 6 |
| chr7_-_109616548 | 0.07 |
ENSMUST00000077909.1
ENSMUST00000084738.3 |
St5
|
suppression of tumorigenicity 5 |
| chr4_-_143299463 | 0.06 |
ENSMUST00000119654.1
|
Pdpn
|
podoplanin |
| chr1_+_34005872 | 0.05 |
ENSMUST00000182296.1
|
Dst
|
dystonin |
| chr2_-_162661075 | 0.05 |
ENSMUST00000109442.1
ENSMUST00000109445.2 ENSMUST00000109443.1 ENSMUST00000109441.1 |
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr11_+_105126425 | 0.05 |
ENSMUST00000021030.7
|
Mettl2
|
methyltransferase like 2 |
| chr2_+_25289899 | 0.04 |
ENSMUST00000028337.6
|
Lrrc26
|
leucine rich repeat containing 26 |
| chrX_-_167855061 | 0.04 |
ENSMUST00000112146.1
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr10_+_102159000 | 0.03 |
ENSMUST00000020039.6
|
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pgr_Nr3c1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 13.9 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.9 | 3.7 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.8 | 2.4 | GO:0006507 | GPI anchor release(GO:0006507) |
| 0.6 | 1.7 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.5 | 5.4 | GO:0006569 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.4 | 1.3 | GO:0072034 | intrahepatic bile duct development(GO:0035622) renal vesicle induction(GO:0072034) |
| 0.3 | 3.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 3.3 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.3 | 1.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.3 | 1.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.3 | 13.1 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.3 | 0.8 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.3 | 1.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 1.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 1.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 0.4 | GO:0003169 | coronary vein morphogenesis(GO:0003169) |
| 0.2 | 2.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.2 | 1.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.2 | 1.4 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.2 | 1.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.2 | 1.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 0.5 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.2 | 1.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 1.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.9 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.4 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.1 | 0.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.6 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) carbon catabolite activation of transcription(GO:0045991) |
| 0.1 | 0.3 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.4 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.1 | 0.2 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 3.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.6 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.3 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 1.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 0.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.3 | GO:0072053 | renal inner medulla development(GO:0072053) |
| 0.1 | 1.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 1.4 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 2.0 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 2.5 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.6 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.0 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 1.9 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 1.1 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.9 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.6 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.0 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.0 | 0.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 4.5 | GO:0015833 | peptide transport(GO:0015833) |
| 0.0 | 1.8 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 2.6 | GO:0015711 | organic anion transport(GO:0015711) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.2 | 2.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 3.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 3.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 1.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.3 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 1.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 1.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 12.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.7 | 2.9 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 1.8 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.3 | 1.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 2.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 2.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 3.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 0.6 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.2 | 1.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 1.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 1.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 1.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.4 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.9 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.4 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 13.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.8 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.9 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.4 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.1 | 0.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 1.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.8 | GO:0030169 | signaling pattern recognition receptor activity(GO:0008329) low-density lipoprotein particle binding(GO:0030169) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 1.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 5.4 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 1.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.4 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 1.2 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 7.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 1.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.5 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.9 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 3.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 5.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 2.9 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 2.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 3.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.8 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |