Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Zfp691
Z-value: 0.96
Transcription factors associated with Zfp691
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp691
|
ENSMUSG00000045268.7 | zinc finger protein 691 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp691 | mm10_v2_chr4_-_119173849_119173901 | -0.47 | 4.2e-03 | Click! |
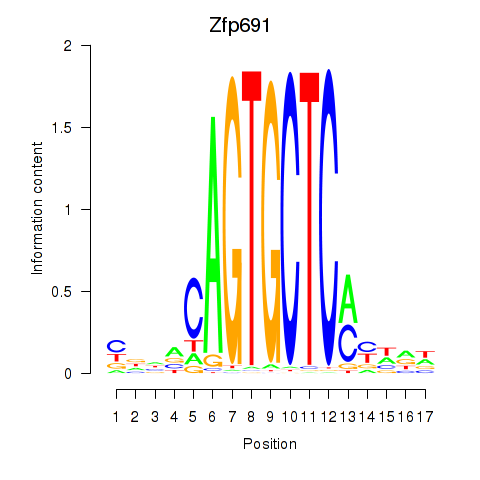
Activity profile of Zfp691 motif
Sorted Z-values of Zfp691 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_51853699 | 3.24 |
ENSMUST00000169070.1
ENSMUST00000074477.6 |
Ear6
|
eosinophil-associated, ribonuclease A family, member 6 |
| chr4_-_118457450 | 2.74 |
ENSMUST00000106375.1
ENSMUST00000006556.3 ENSMUST00000168404.1 |
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr8_+_21776567 | 2.56 |
ENSMUST00000051017.8
|
Defb1
|
defensin beta 1 |
| chr6_+_41302265 | 2.51 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr4_-_118457509 | 2.49 |
ENSMUST00000102671.3
|
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr2_-_121037048 | 2.02 |
ENSMUST00000102490.3
|
Epb4.2
|
erythrocyte protein band 4.2 |
| chr13_-_100786402 | 1.97 |
ENSMUST00000174038.1
ENSMUST00000091295.7 ENSMUST00000072119.8 |
Ccnb1
|
cyclin B1 |
| chr9_+_62858085 | 1.90 |
ENSMUST00000034777.6
ENSMUST00000163820.1 |
Calml4
|
calmodulin-like 4 |
| chr11_-_99493112 | 1.69 |
ENSMUST00000006969.7
|
Krt23
|
keratin 23 |
| chr11_-_121039400 | 1.66 |
ENSMUST00000026159.5
|
Cd7
|
CD7 antigen |
| chr3_+_98222148 | 1.66 |
ENSMUST00000029469.4
|
Reg4
|
regenerating islet-derived family, member 4 |
| chr12_+_102129019 | 1.49 |
ENSMUST00000079020.4
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr17_-_23684019 | 1.38 |
ENSMUST00000085989.5
|
Cldn9
|
claudin 9 |
| chr16_+_32608920 | 1.32 |
ENSMUST00000023486.8
|
Tfrc
|
transferrin receptor |
| chrX_+_136666375 | 1.32 |
ENSMUST00000060904.4
ENSMUST00000113100.1 ENSMUST00000128040.1 |
Tceal3
|
transcription elongation factor A (SII)-like 3 |
| chr3_-_15332285 | 1.30 |
ENSMUST00000108361.1
|
Gm9733
|
predicted gene 9733 |
| chr7_+_28863831 | 1.18 |
ENSMUST00000138272.1
|
Lgals7
|
lectin, galactose binding, soluble 7 |
| chr19_-_11266122 | 1.17 |
ENSMUST00000169159.1
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr3_-_15848419 | 1.09 |
ENSMUST00000108354.1
ENSMUST00000108349.1 ENSMUST00000108352.2 ENSMUST00000108350.1 ENSMUST00000050623.4 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr6_-_113633127 | 1.01 |
ENSMUST00000180964.1
|
1700015O11Rik
|
RIKEN cDNA 1700015O11 gene |
| chr17_-_25754327 | 0.86 |
ENSMUST00000075884.6
|
Msln
|
mesothelin |
| chr13_+_53525703 | 0.82 |
ENSMUST00000081132.4
|
Gm5449
|
predicted pseudogene 5449 |
| chr6_-_128788486 | 0.80 |
ENSMUST00000172601.1
|
Klrb1c
|
killer cell lectin-like receptor subfamily B member 1C |
| chr13_-_55528511 | 0.75 |
ENSMUST00000047877.4
|
Dok3
|
docking protein 3 |
| chr12_+_86678685 | 0.72 |
ENSMUST00000021681.3
|
Vash1
|
vasohibin 1 |
| chr11_-_79523760 | 0.72 |
ENSMUST00000179322.1
|
Evi2b
|
ecotropic viral integration site 2b |
| chr5_+_37050854 | 0.67 |
ENSMUST00000043794.4
|
Jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr2_-_10130638 | 0.67 |
ENSMUST00000042290.7
|
Itih2
|
inter-alpha trypsin inhibitor, heavy chain 2 |
| chr10_-_86732409 | 0.63 |
ENSMUST00000070435.4
|
Fabp3-ps1
|
fatty acid binding protein 3, muscle and heart, pseudogene 1 |
| chr9_-_103761820 | 0.62 |
ENSMUST00000049452.8
|
Tmem108
|
transmembrane protein 108 |
| chr8_+_69808672 | 0.61 |
ENSMUST00000036074.8
ENSMUST00000123453.1 |
Gmip
|
Gem-interacting protein |
| chr5_+_35893319 | 0.58 |
ENSMUST00000064571.4
|
Afap1
|
actin filament associated protein 1 |
| chr19_+_11516473 | 0.58 |
ENSMUST00000163078.1
|
Ms4a6b
|
membrane-spanning 4-domains, subfamily A, member 6B |
| chr2_+_181715005 | 0.58 |
ENSMUST00000071585.3
ENSMUST00000148334.1 ENSMUST00000108763.1 |
Oprl1
|
opioid receptor-like 1 |
| chr1_+_134193432 | 0.57 |
ENSMUST00000038445.6
|
Mybph
|
myosin binding protein H |
| chr2_+_119112793 | 0.56 |
ENSMUST00000140939.1
ENSMUST00000028795.3 |
Rad51
|
RAD51 homolog |
| chr17_-_32800938 | 0.56 |
ENSMUST00000080905.6
|
Zfp811
|
zinc finger protein 811 |
| chr3_+_98280427 | 0.55 |
ENSMUST00000090746.2
ENSMUST00000120541.1 |
Hmgcs2
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 |
| chr16_+_90386382 | 0.55 |
ENSMUST00000065856.6
|
Hunk
|
hormonally upregulated Neu-associated kinase |
| chr16_+_44811733 | 0.55 |
ENSMUST00000176819.1
ENSMUST00000176321.1 |
Cd200r4
|
CD200 receptor 4 |
| chr10_+_115569986 | 0.53 |
ENSMUST00000173620.1
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr11_+_5569679 | 0.52 |
ENSMUST00000109856.1
ENSMUST00000109855.1 ENSMUST00000118112.2 |
Ankrd36
|
ankyrin repeat domain 36 |
| chr4_-_117765550 | 0.52 |
ENSMUST00000062747.5
|
Klf17
|
Kruppel-like factor 17 |
| chr11_-_116694802 | 0.50 |
ENSMUST00000079545.5
|
St6galnac2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr16_+_44765732 | 0.47 |
ENSMUST00000057488.8
|
Cd200r1
|
CD200 receptor 1 |
| chr7_+_64153835 | 0.47 |
ENSMUST00000085222.5
ENSMUST00000107525.1 |
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr6_-_87533219 | 0.46 |
ENSMUST00000113637.2
ENSMUST00000071024.6 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr4_-_141416002 | 0.45 |
ENSMUST00000006378.2
ENSMUST00000105788.1 |
Clcnkb
|
chloride channel Kb |
| chr2_-_170194033 | 0.45 |
ENSMUST00000180625.1
|
Gm17619
|
predicted gene, 17619 |
| chr10_-_88356990 | 0.44 |
ENSMUST00000020249.1
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr11_-_121118677 | 0.42 |
ENSMUST00000039146.3
|
Tex19.2
|
testis expressed gene 19.2 |
| chr8_+_72319033 | 0.41 |
ENSMUST00000067912.7
|
Klf2
|
Kruppel-like factor 2 (lung) |
| chrX_-_101268218 | 0.41 |
ENSMUST00000033664.7
|
Il2rg
|
interleukin 2 receptor, gamma chain |
| chr16_-_32165454 | 0.40 |
ENSMUST00000115163.3
ENSMUST00000144345.1 ENSMUST00000143682.1 ENSMUST00000115165.3 ENSMUST00000099991.4 ENSMUST00000130410.1 |
Nrros
|
negative regulator of reactive oxygen species |
| chr9_-_58313189 | 0.40 |
ENSMUST00000061799.8
|
Loxl1
|
lysyl oxidase-like 1 |
| chr17_-_65772686 | 0.38 |
ENSMUST00000070673.7
|
Rab31
|
RAB31, member RAS oncogene family |
| chr9_-_108452377 | 0.37 |
ENSMUST00000035232.7
|
Klhdc8b
|
kelch domain containing 8B |
| chr9_+_124121534 | 0.37 |
ENSMUST00000111442.1
ENSMUST00000171499.2 |
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr13_-_60936550 | 0.36 |
ENSMUST00000021880.9
|
Ctla2a
|
cytotoxic T lymphocyte-associated protein 2 alpha |
| chr13_-_60897439 | 0.36 |
ENSMUST00000171347.1
ENSMUST00000021884.8 |
Ctla2b
|
cytotoxic T lymphocyte-associated protein 2 beta |
| chr3_-_15575065 | 0.34 |
ENSMUST00000091319.4
|
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr10_-_112928974 | 0.34 |
ENSMUST00000099276.2
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr7_+_143069249 | 0.33 |
ENSMUST00000060433.3
ENSMUST00000133410.2 ENSMUST00000105920.1 ENSMUST00000177841.1 ENSMUST00000147995.1 ENSMUST00000137856.1 |
Tssc4
|
tumor-suppressing subchromosomal transferable fragment 4 |
| chr8_+_72135247 | 0.32 |
ENSMUST00000003575.9
|
Tpm4
|
tropomyosin 4 |
| chr7_+_118633729 | 0.31 |
ENSMUST00000057320.7
|
Tmc5
|
transmembrane channel-like gene family 5 |
| chr7_-_104390586 | 0.31 |
ENSMUST00000106828.1
|
Trim30c
|
tripartite motif-containing 30C |
| chr8_-_124569696 | 0.30 |
ENSMUST00000063278.6
|
Agt
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| chr4_+_32657107 | 0.30 |
ENSMUST00000071642.4
ENSMUST00000178134.1 |
Mdn1
|
midasin homolog (yeast) |
| chr9_+_32696005 | 0.30 |
ENSMUST00000034534.6
ENSMUST00000050797.7 ENSMUST00000184887.1 |
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr5_+_128601106 | 0.30 |
ENSMUST00000117102.2
|
Fzd10
|
frizzled homolog 10 (Drosophila) |
| chr11_-_94973447 | 0.29 |
ENSMUST00000100551.4
ENSMUST00000152042.1 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chr1_-_176807124 | 0.27 |
ENSMUST00000057037.7
|
Cep170
|
centrosomal protein 170 |
| chr14_-_70323783 | 0.26 |
ENSMUST00000151011.1
|
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr1_+_21272338 | 0.25 |
ENSMUST00000178379.1
|
Gm7094
|
predicted pseudogene 7094 |
| chr3_-_108889990 | 0.25 |
ENSMUST00000053065.4
ENSMUST00000102620.3 |
Fndc7
|
fibronectin type III domain containing 7 |
| chr15_+_101411053 | 0.25 |
ENSMUST00000147662.1
|
Krt7
|
keratin 7 |
| chr6_+_70844499 | 0.25 |
ENSMUST00000034093.8
ENSMUST00000162950.1 |
Eif2ak3
|
eukaryotic translation initiation factor 2 alpha kinase 3 |
| chr8_-_77517898 | 0.24 |
ENSMUST00000076316.4
|
Arhgap10
|
Rho GTPase activating protein 10 |
| chr2_-_131352857 | 0.23 |
ENSMUST00000059372.4
|
Rnf24
|
ring finger protein 24 |
| chr10_+_94576254 | 0.20 |
ENSMUST00000117929.1
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr3_-_82074639 | 0.20 |
ENSMUST00000029635.8
|
Gucy1b3
|
guanylate cyclase 1, soluble, beta 3 |
| chr4_+_132535542 | 0.19 |
ENSMUST00000094657.3
ENSMUST00000105940.3 ENSMUST00000105939.3 ENSMUST00000150207.1 |
Dnajc8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr4_+_124714776 | 0.19 |
ENSMUST00000030734.4
|
Sf3a3
|
splicing factor 3a, subunit 3 |
| chr7_+_64153916 | 0.18 |
ENSMUST00000107527.2
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr15_+_82252397 | 0.17 |
ENSMUST00000136948.1
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr11_-_101175440 | 0.16 |
ENSMUST00000062759.3
|
Ccr10
|
chemokine (C-C motif) receptor 10 |
| chr14_+_73143046 | 0.16 |
ENSMUST00000170677.1
ENSMUST00000167401.1 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr2_-_180709985 | 0.16 |
ENSMUST00000103057.1
ENSMUST00000103055.1 |
Dido1
|
death inducer-obliterator 1 |
| chr6_+_145934113 | 0.16 |
ENSMUST00000032383.7
|
Sspn
|
sarcospan |
| chr19_-_11660516 | 0.16 |
ENSMUST00000135994.1
ENSMUST00000121793.1 ENSMUST00000069681.3 |
Plac1l
|
placenta-specific 1-like |
| chr8_+_88521344 | 0.15 |
ENSMUST00000034086.5
|
Nkd1
|
naked cuticle 1 homolog (Drosophila) |
| chr14_+_73142863 | 0.15 |
ENSMUST00000171767.1
ENSMUST00000163533.1 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr2_+_24962400 | 0.15 |
ENSMUST00000028351.3
|
Dph7
|
diphthamine biosynethesis 7 |
| chr6_-_141773810 | 0.14 |
ENSMUST00000148411.1
|
Gm5724
|
predicted gene 5724 |
| chr8_-_22805596 | 0.14 |
ENSMUST00000163739.1
|
Ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chrX_-_95478107 | 0.14 |
ENSMUST00000033549.2
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr11_-_116454347 | 0.13 |
ENSMUST00000093909.4
|
Qrich2
|
glutamine rich 2 |
| chr7_-_143685863 | 0.13 |
ENSMUST00000152703.1
|
Tnfrsf23
|
tumor necrosis factor receptor superfamily, member 23 |
| chr13_-_68582087 | 0.12 |
ENSMUST00000045827.4
|
Mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr1_+_51289106 | 0.12 |
ENSMUST00000051572.6
|
Sdpr
|
serum deprivation response |
| chr3_-_108889706 | 0.11 |
ENSMUST00000180063.1
|
Fndc7
|
fibronectin type III domain containing 7 |
| chr7_+_140093388 | 0.09 |
ENSMUST00000026540.8
|
Prap1
|
proline-rich acidic protein 1 |
| chr2_+_26591423 | 0.09 |
ENSMUST00000152988.2
ENSMUST00000149789.1 |
Egfl7
|
EGF-like domain 7 |
| chr12_+_116077720 | 0.08 |
ENSMUST00000011315.3
|
Vipr2
|
vasoactive intestinal peptide receptor 2 |
| chr11_-_49114874 | 0.08 |
ENSMUST00000109201.1
|
Olfr1396
|
olfactory receptor 1396 |
| chr7_-_38019505 | 0.08 |
ENSMUST00000085513.4
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr2_+_91265252 | 0.08 |
ENSMUST00000028691.6
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr13_-_54468805 | 0.08 |
ENSMUST00000026990.5
|
Thoc3
|
THO complex 3 |
| chr10_-_60003056 | 0.06 |
ENSMUST00000182912.1
ENSMUST00000020307.4 ENSMUST00000182898.1 |
Anapc16
|
anaphase promoting complex subunit 16 |
| chr9_-_58202281 | 0.06 |
ENSMUST00000163897.1
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr15_+_97784355 | 0.06 |
ENSMUST00000117892.1
|
Slc48a1
|
solute carrier family 48 (heme transporter), member 1 |
| chr7_-_143649614 | 0.05 |
ENSMUST00000129476.1
ENSMUST00000084396.3 ENSMUST00000075588.6 ENSMUST00000146692.1 |
Tnfrsf22
|
tumor necrosis factor receptor superfamily, member 22 |
| chr9_+_27299205 | 0.05 |
ENSMUST00000115247.1
ENSMUST00000133213.1 |
Igsf9b
|
immunoglobulin superfamily, member 9B |
| chr9_-_106887000 | 0.05 |
ENSMUST00000055843.7
|
Rbm15b
|
RNA binding motif protein 15B |
| chr10_-_77166545 | 0.05 |
ENSMUST00000081654.6
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr11_+_120467635 | 0.05 |
ENSMUST00000140862.1
ENSMUST00000106205.1 ENSMUST00000106203.1 ENSMUST00000026900.7 |
Hgs
|
HGF-regulated tyrosine kinase substrate |
| chr9_-_104262900 | 0.05 |
ENSMUST00000035170.6
|
Dnajc13
|
DnaJ (Hsp40) homolog, subfamily C, member 13 |
| chr7_-_12422751 | 0.04 |
ENSMUST00000080348.5
|
Zfp551
|
zinc fingr protein 551 |
| chr14_+_73142490 | 0.04 |
ENSMUST00000170370.1
ENSMUST00000164822.1 ENSMUST00000165429.1 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr10_+_60003321 | 0.04 |
ENSMUST00000164083.2
|
Ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr10_+_4482502 | 0.03 |
ENSMUST00000138112.1
|
Ccdc170
|
coiled-coil domain containing 170 |
| chr4_-_129623870 | 0.03 |
ENSMUST00000106035.1
ENSMUST00000150357.1 ENSMUST00000030586.8 |
Ccdc28b
|
coiled coil domain containing 28B |
| chr14_+_73142591 | 0.02 |
ENSMUST00000170368.1
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr9_-_50617428 | 0.02 |
ENSMUST00000131351.1
ENSMUST00000171462.1 |
AU019823
|
expressed sequence AU019823 |
| chr2_-_106002008 | 0.01 |
ENSMUST00000155811.1
|
Dnajc24
|
DnaJ (Hsp40) homolog, subfamily C, member 24 |
| chr9_-_50603792 | 0.01 |
ENSMUST00000000175.4
|
Sdhd
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp691
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 0.7 | 2.0 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.2 | 0.6 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.7 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.6 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.4 | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.1 | 0.3 | GO:0003330 | regulation of renal output by angiotensin(GO:0002019) regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) L-lysine transport(GO:1902022) |
| 0.1 | 0.5 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 1.3 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.4 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.6 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.4 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.1 | 0.2 | GO:0099548 | trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 1.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.5 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.3 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 1.7 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.8 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.7 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 2.5 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.6 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.1 | GO:0033353 | sulfur amino acid catabolic process(GO:0000098) S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.2 | 0.7 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 1.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.6 | GO:0005859 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 0.0 | 1.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.4 | 1.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.2 | 1.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.4 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 5.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 3.2 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.5 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 2.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 5.2 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 1.3 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |