Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Aire
Z-value: 0.72
Transcription factors associated with Aire
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Aire
|
ENSMUSG00000000731.16 | autoimmune regulator (autoimmune polyendocrinopathy candidiasis ectodermal dystrophy) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Aire | mm39_v1_chr10_-_77879414_77879445 | -0.18 | 3.1e-01 | Click! |
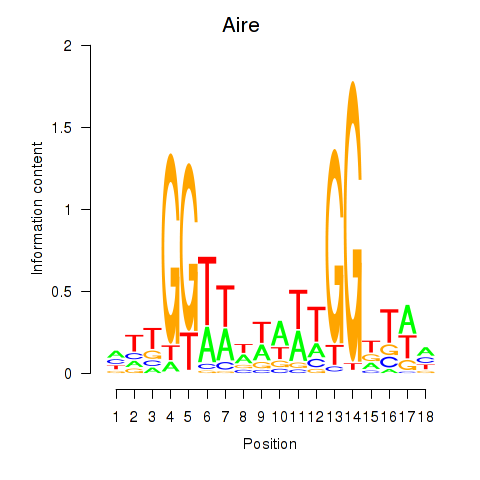
Activity profile of Aire motif
Sorted Z-values of Aire motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39275518 | 4.05 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr6_+_42222841 | 3.51 |
ENSMUST00000031897.8
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr10_-_95678786 | 2.89 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr10_-_95678748 | 2.81 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543 |
| chr5_+_8010445 | 2.67 |
ENSMUST00000115421.3
|
Steap4
|
STEAP family member 4 |
| chr1_+_88128323 | 2.33 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr12_+_84098888 | 1.95 |
ENSMUST00000120927.8
ENSMUST00000021653.8 |
Acot3
|
acyl-CoA thioesterase 3 |
| chr5_+_130398261 | 1.76 |
ENSMUST00000086029.10
|
Caln1
|
calneuron 1 |
| chrM_+_11735 | 1.74 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr5_+_31079177 | 1.67 |
ENSMUST00000031053.15
ENSMUST00000202752.2 |
Khk
|
ketohexokinase |
| chr5_+_137979763 | 1.61 |
ENSMUST00000035390.7
|
Azgp1
|
alpha-2-glycoprotein 1, zinc |
| chr7_-_127494750 | 1.53 |
ENSMUST00000033074.8
|
Vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr7_-_126275529 | 1.47 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr15_+_10358611 | 1.46 |
ENSMUST00000110541.8
ENSMUST00000110542.8 |
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chrM_+_7758 | 1.45 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr16_+_22877000 | 1.41 |
ENSMUST00000039492.14
ENSMUST00000023589.15 ENSMUST00000089902.8 |
Kng1
|
kininogen 1 |
| chr15_+_10358643 | 1.40 |
ENSMUST00000022858.8
|
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr5_-_121798541 | 1.28 |
ENSMUST00000031412.12
ENSMUST00000111770.2 |
Acad10
|
acyl-Coenzyme A dehydrogenase family, member 10 |
| chr15_+_10981833 | 1.24 |
ENSMUST00000070877.7
|
Amacr
|
alpha-methylacyl-CoA racemase |
| chr11_-_69586884 | 1.21 |
ENSMUST00000180587.8
|
Tnfsfm13
|
tumor necrosis factor (ligand) superfamily, membrane-bound member 13 |
| chr16_-_18904240 | 1.19 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr10_-_106959444 | 1.18 |
ENSMUST00000165067.9
|
Acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chrM_+_7006 | 1.18 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr9_-_108140925 | 1.17 |
ENSMUST00000171412.7
ENSMUST00000195429.6 ENSMUST00000080435.9 |
Dag1
|
dystroglycan 1 |
| chr16_-_22847808 | 1.10 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chr16_-_22847829 | 1.09 |
ENSMUST00000100046.9
|
Kng2
|
kininogen 2 |
| chr15_-_89263790 | 1.07 |
ENSMUST00000238996.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chrM_+_9459 | 1.07 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chrM_+_7779 | 1.02 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr14_+_3575406 | 1.01 |
ENSMUST00000124353.2
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr17_+_56935118 | 1.00 |
ENSMUST00000112979.4
|
Catsperd
|
cation channel sperm associated auxiliary subunit delta |
| chrM_+_14138 | 0.97 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr5_-_105491795 | 0.93 |
ENSMUST00000171587.2
|
Gbp11
|
guanylate binding protein 11 |
| chrM_-_14061 | 0.92 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr12_+_37292029 | 0.90 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chrM_+_8603 | 0.85 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr16_-_19079594 | 0.85 |
ENSMUST00000103752.3
ENSMUST00000197518.2 |
Iglv2
|
immunoglobulin lambda variable 2 |
| chr17_-_74257164 | 0.83 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr3_+_57332735 | 0.82 |
ENSMUST00000029377.8
|
Tm4sf4
|
transmembrane 4 superfamily member 4 |
| chr1_-_128287347 | 0.79 |
ENSMUST00000190495.2
ENSMUST00000027601.11 |
Mcm6
|
minichromosome maintenance complex component 6 |
| chr7_-_126522014 | 0.77 |
ENSMUST00000134134.3
ENSMUST00000119781.8 ENSMUST00000121612.4 |
Tmem219
|
transmembrane protein 219 |
| chr13_+_64309675 | 0.71 |
ENSMUST00000021929.10
|
Habp4
|
hyaluronic acid binding protein 4 |
| chr10_-_127587576 | 0.71 |
ENSMUST00000079692.6
|
Gpr182
|
G protein-coupled receptor 182 |
| chr1_-_90771674 | 0.61 |
ENSMUST00000097653.11
ENSMUST00000188587.7 ENSMUST00000056925.16 ENSMUST00000187753.2 |
Col6a3
|
collagen, type VI, alpha 3 |
| chr7_-_19398930 | 0.57 |
ENSMUST00000055242.11
|
Clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr17_-_59320257 | 0.57 |
ENSMUST00000174122.2
ENSMUST00000025065.12 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr7_-_45136235 | 0.55 |
ENSMUST00000210701.2
|
Gm45808
|
predicted gene 45808 |
| chr3_-_89319088 | 0.55 |
ENSMUST00000107429.10
ENSMUST00000129308.9 ENSMUST00000107426.8 ENSMUST00000050398.11 ENSMUST00000162701.2 |
Flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr17_-_35827676 | 0.54 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr14_+_76714350 | 0.48 |
ENSMUST00000140251.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr17_+_31652029 | 0.48 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr9_+_45924120 | 0.44 |
ENSMUST00000120463.9
ENSMUST00000120247.8 |
Sik3
|
SIK family kinase 3 |
| chr11_+_97206542 | 0.43 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr9_+_108883907 | 0.43 |
ENSMUST00000154184.5
|
Shisa5
|
shisa family member 5 |
| chr11_+_48977495 | 0.42 |
ENSMUST00000152914.2
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr1_-_45542442 | 0.39 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr5_-_100521343 | 0.38 |
ENSMUST00000182433.8
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr9_+_45924105 | 0.37 |
ENSMUST00000126865.8
|
Sik3
|
SIK family kinase 3 |
| chr11_-_66943389 | 0.36 |
ENSMUST00000116363.2
|
Adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese dependent |
| chr1_-_65225617 | 0.31 |
ENSMUST00000186222.7
ENSMUST00000169032.8 ENSMUST00000191459.2 ENSMUST00000188876.7 |
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr7_+_45546365 | 0.31 |
ENSMUST00000069772.16
ENSMUST00000210503.2 ENSMUST00000209350.2 |
Tmem143
|
transmembrane protein 143 |
| chr17_+_35455532 | 0.31 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr5_-_50216249 | 0.29 |
ENSMUST00000030971.7
|
Adgra3
|
adhesion G protein-coupled receptor A3 |
| chr1_-_65225572 | 0.29 |
ENSMUST00000188109.7
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr5_+_115034997 | 0.29 |
ENSMUST00000031542.13
ENSMUST00000146072.8 ENSMUST00000150361.2 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr17_-_56343531 | 0.28 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr11_+_17161912 | 0.25 |
ENSMUST00000046955.7
|
Wdr92
|
WD repeat domain 92 |
| chr15_-_76809607 | 0.25 |
ENSMUST00000229865.2
ENSMUST00000229055.2 |
Zfp647
|
zinc finger protein 647 |
| chr12_+_36207113 | 0.23 |
ENSMUST00000041640.5
|
Ankmy2
|
ankyrin repeat and MYND domain containing 2 |
| chr6_-_3399451 | 0.22 |
ENSMUST00000120087.6
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr6_+_91678630 | 0.22 |
ENSMUST00000205828.2
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr6_-_91492860 | 0.22 |
ENSMUST00000206476.2
ENSMUST00000032182.5 |
Xpc
|
xeroderma pigmentosum, complementation group C |
| chr1_-_90771638 | 0.21 |
ENSMUST00000130846.9
|
Col6a3
|
collagen, type VI, alpha 3 |
| chr7_+_28869770 | 0.20 |
ENSMUST00000033886.8
ENSMUST00000209019.2 ENSMUST00000208330.2 |
Ggn
|
gametogenetin |
| chr11_+_66943453 | 0.19 |
ENSMUST00000108690.10
ENSMUST00000092996.5 |
Sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr2_+_89804937 | 0.18 |
ENSMUST00000214630.2
ENSMUST00000111512.10 ENSMUST00000144710.3 ENSMUST00000216678.2 |
Olfr1260
|
olfactory receptor 1260 |
| chr18_+_37858753 | 0.18 |
ENSMUST00000066149.9
|
Pcdhga8
|
protocadherin gamma subfamily A, 8 |
| chr8_+_112370088 | 0.17 |
ENSMUST00000077791.8
ENSMUST00000211926.2 |
Zfp1
|
zinc finger protein 1 |
| chr3_+_89986831 | 0.17 |
ENSMUST00000029549.16
|
Tpm3
|
tropomyosin 3, gamma |
| chr11_+_105866030 | 0.15 |
ENSMUST00000001964.8
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr1_+_44159106 | 0.15 |
ENSMUST00000114709.3
ENSMUST00000129068.2 |
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr8_+_112370033 | 0.15 |
ENSMUST00000212072.2
|
Zfp1
|
zinc finger protein 1 |
| chr7_+_30681287 | 0.13 |
ENSMUST00000128384.3
|
Fam187b
|
family with sequence similarity 187, member B |
| chr13_+_49658249 | 0.12 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr6_+_41039255 | 0.12 |
ENSMUST00000103266.3
|
Trbv5
|
T cell receptor beta, variable 5 |
| chr9_-_19163273 | 0.11 |
ENSMUST00000214019.2
ENSMUST00000214267.2 |
Olfr843
|
olfactory receptor 843 |
| chr10_+_3822667 | 0.11 |
ENSMUST00000136671.8
ENSMUST00000042438.13 |
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr3_+_92339886 | 0.11 |
ENSMUST00000067102.3
|
Sprr2k
|
small proline-rich protein 2K |
| chrY_-_79161056 | 0.10 |
ENSMUST00000179922.2
|
Gm20917
|
predicted gene, 20917 |
| chr11_-_49077864 | 0.10 |
ENSMUST00000153999.3
ENSMUST00000066531.13 |
Btnl9
|
butyrophilin-like 9 |
| chr11_-_115518774 | 0.09 |
ENSMUST00000154623.2
ENSMUST00000106503.10 ENSMUST00000141614.3 |
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr2_-_93841152 | 0.09 |
ENSMUST00000111240.8
|
Alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr8_+_65399831 | 0.09 |
ENSMUST00000026595.13
ENSMUST00000209852.2 ENSMUST00000079896.9 |
Tmem192
|
transmembrane protein 192 |
| chr3_+_82933383 | 0.09 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chr2_-_93841062 | 0.09 |
ENSMUST00000040005.13
ENSMUST00000126378.8 |
Alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr16_+_36695479 | 0.08 |
ENSMUST00000023534.7
ENSMUST00000114812.9 ENSMUST00000134616.8 |
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1 |
| chrX_-_91059789 | 0.08 |
ENSMUST00000099471.3
ENSMUST00000072269.2 |
Mageb1
|
MAGE family member B1 |
| chr9_-_110818679 | 0.08 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chr16_-_91415873 | 0.07 |
ENSMUST00000143058.2
ENSMUST00000049244.10 ENSMUST00000169982.2 ENSMUST00000133731.2 |
Dnajc28
|
DnaJ heat shock protein family (Hsp40) member C28 |
| chr14_-_50919102 | 0.07 |
ENSMUST00000078075.5
|
Olfr747
|
olfactory receptor 747 |
| chr4_-_118667141 | 0.07 |
ENSMUST00000084313.5
ENSMUST00000219094.2 |
Olfr1335
|
olfactory receptor 1335 |
| chr2_-_112089627 | 0.07 |
ENSMUST00000043970.2
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr11_-_96002784 | 0.06 |
ENSMUST00000097162.6
ENSMUST00000068686.13 |
Calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr10_-_130116088 | 0.06 |
ENSMUST00000061571.5
|
Neurod4
|
neurogenic differentiation 4 |
| chr2_+_83554741 | 0.06 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr2_+_128433125 | 0.06 |
ENSMUST00000155430.8
|
Spdye4c
|
speedy/RINGO cell cycle regulator family, member E4C |
| chr3_-_92734546 | 0.05 |
ENSMUST00000072363.5
|
Kprp
|
keratinocyte expressed, proline-rich |
| chr1_-_85247864 | 0.05 |
ENSMUST00000159582.8
ENSMUST00000161267.8 |
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr1_+_189460461 | 0.05 |
ENSMUST00000097442.9
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr16_-_36695166 | 0.05 |
ENSMUST00000075946.12
|
Eaf2
|
ELL associated factor 2 |
| chr10_+_80441855 | 0.04 |
ENSMUST00000178231.2
|
Gm49322
|
predicted gene, 49322 |
| chr3_-_95125190 | 0.04 |
ENSMUST00000136139.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr8_+_47439916 | 0.04 |
ENSMUST00000039840.15
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr15_-_98063441 | 0.04 |
ENSMUST00000123922.2
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr17_+_87943401 | 0.03 |
ENSMUST00000235125.2
ENSMUST00000053577.9 ENSMUST00000234009.2 |
Epcam
|
epithelial cell adhesion molecule |
| chr15_-_75793312 | 0.03 |
ENSMUST00000053918.9
|
Pycrl
|
pyrroline-5-carboxylate reductase-like |
| chrX_-_6907858 | 0.02 |
ENSMUST00000115752.8
|
Ccnb3
|
cyclin B3 |
| chr6_+_11907808 | 0.02 |
ENSMUST00000155037.4
|
Phf14
|
PHD finger protein 14 |
| chr1_+_173877941 | 0.02 |
ENSMUST00000062665.4
|
Olfr432
|
olfactory receptor 432 |
| chr8_-_110688716 | 0.02 |
ENSMUST00000001722.14
ENSMUST00000051430.7 |
Marveld3
|
MARVEL (membrane-associating) domain containing 3 |
| chr9_-_39920878 | 0.02 |
ENSMUST00000216463.3
|
Olfr980
|
olfactory receptor 980 |
| chr1_+_21419819 | 0.01 |
ENSMUST00000088407.4
|
Khdc1a
|
KH domain containing 1A |
| chr13_-_105429514 | 0.01 |
ENSMUST00000224011.2
|
Rnf180
|
ring finger protein 180 |
| chr14_+_53531798 | 0.01 |
ENSMUST00000103626.3
|
Trav9n-4
|
T cell receptor alpha variable 9N-4 |
| chr5_-_105198913 | 0.01 |
ENSMUST00000112718.5
|
Gbp8
|
guanylate-binding protein 8 |
| chr7_-_84059170 | 0.01 |
ENSMUST00000208995.2
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr3_+_107008867 | 0.01 |
ENSMUST00000038695.6
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr11_+_62349238 | 0.00 |
ENSMUST00000014389.6
|
Pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
Network of associatons between targets according to the STRING database.
First level regulatory network of Aire
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0042853 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) |
| 0.7 | 2.0 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.4 | 1.7 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.4 | 1.2 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.4 | 1.5 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.3 | 2.7 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 2.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.5 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.2 | 0.8 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.2 | 0.6 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 1.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 4.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 1.0 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 1.2 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.6 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.1 | 1.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 2.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.2 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.1 | 0.2 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.9 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 3.7 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 1.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 | 2.9 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 3.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.9 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:0035553 | oxidative single-stranded DNA demethylation(GO:0035552) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 1.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.0 | 1.1 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.7 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.8 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.6 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.2 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 1.0 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 1.0 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.1 | 3.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 7.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 3.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.6 | 1.7 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.5 | 1.5 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.5 | 2.9 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.4 | 1.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 2.7 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 0.9 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.2 | 1.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 1.0 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 0.8 | GO:0043546 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 3.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 0.6 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 1.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.0 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 3.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.0 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 2.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.7 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 0.2 | GO:0001761 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.1 | 2.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.1 | 1.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) bradykinin receptor binding(GO:0031711) |
| 0.0 | 2.8 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 0.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |