Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for E2f4
Z-value: 3.82
Transcription factors associated with E2f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f4
|
ENSMUSG00000014859.10 | E2F transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f4 | mm39_v1_chr8_+_106024294_106024367 | 0.97 | 2.0e-22 | Click! |
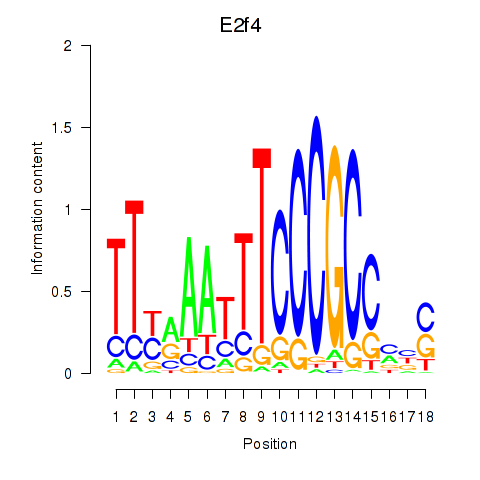
Activity profile of E2f4 motif
Sorted Z-values of E2f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_37806912 | 41.79 |
ENSMUST00000108023.10
|
Ccne1
|
cyclin E1 |
| chr6_+_124807176 | 40.50 |
ENSMUST00000131847.8
ENSMUST00000151674.3 |
Cdca3
|
cell division cycle associated 3 |
| chr11_+_68936457 | 39.39 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chr16_-_30129591 | 36.01 |
ENSMUST00000061190.8
|
Gp5
|
glycoprotein 5 (platelet) |
| chr4_+_134238310 | 35.21 |
ENSMUST00000105866.3
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr15_+_79776597 | 33.91 |
ENSMUST00000175714.8
ENSMUST00000109620.10 ENSMUST00000165537.8 ENSMUST00000175752.8 ENSMUST00000176325.8 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr4_-_133694607 | 32.30 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr19_+_6135013 | 30.70 |
ENSMUST00000025704.3
|
Cdca5
|
cell division cycle associated 5 |
| chr18_-_34884555 | 30.62 |
ENSMUST00000060710.9
|
Cdc25c
|
cell division cycle 25C |
| chr12_+_24758240 | 29.84 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr2_+_162896602 | 28.83 |
ENSMUST00000018005.10
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr4_-_133694543 | 28.75 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr11_+_74510413 | 28.68 |
ENSMUST00000100866.3
|
Ccdc92b
|
coiled-coil domain containing 92B |
| chr9_+_72345801 | 28.64 |
ENSMUST00000184604.8
ENSMUST00000034746.10 |
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr1_-_169358912 | 25.22 |
ENSMUST00000192248.2
ENSMUST00000028000.13 |
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr9_+_44245981 | 24.72 |
ENSMUST00000052686.4
|
H2ax
|
H2A.X variant histone |
| chr8_-_54091980 | 24.35 |
ENSMUST00000047768.11
|
Neil3
|
nei like 3 (E. coli) |
| chrX_+_8137372 | 24.18 |
ENSMUST00000127103.8
ENSMUST00000115591.8 |
Slc38a5
|
solute carrier family 38, member 5 |
| chr14_-_87378641 | 23.51 |
ENSMUST00000168889.3
ENSMUST00000022599.14 |
Diaph3
|
diaphanous related formin 3 |
| chr4_-_132072988 | 22.38 |
ENSMUST00000030726.13
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr4_-_133695204 | 22.20 |
ENSMUST00000100472.10
ENSMUST00000136327.2 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr17_-_25946370 | 22.08 |
ENSMUST00000170070.3
ENSMUST00000048054.14 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chrX_+_8137620 | 21.72 |
ENSMUST00000033512.11
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr4_-_132073048 | 21.65 |
ENSMUST00000084250.11
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr9_+_72345267 | 21.49 |
ENSMUST00000183809.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr1_-_33708876 | 20.82 |
ENSMUST00000027312.11
|
Prim2
|
DNA primase, p58 subunit |
| chrX_+_8137881 | 20.17 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr1_-_169359015 | 19.89 |
ENSMUST00000111368.8
|
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr4_-_115980813 | 19.55 |
ENSMUST00000102704.4
ENSMUST00000102705.10 |
Rad54l
|
RAD54 like (S. cerevisiae) |
| chr6_+_113508636 | 18.61 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr18_+_56840813 | 18.23 |
ENSMUST00000025486.9
|
Lmnb1
|
lamin B1 |
| chr9_+_107828136 | 17.34 |
ENSMUST00000049348.9
ENSMUST00000194271.2 |
Traip
|
TRAF-interacting protein |
| chr4_-_133695264 | 17.10 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chrX_+_141464393 | 16.26 |
ENSMUST00000112889.8
ENSMUST00000101198.9 ENSMUST00000112891.8 ENSMUST00000087333.9 |
Tmem164
|
transmembrane protein 164 |
| chr5_+_123214332 | 15.92 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr15_-_76524097 | 15.28 |
ENSMUST00000168185.8
|
Tonsl
|
tonsoku-like, DNA repair protein |
| chr13_-_74085880 | 15.27 |
ENSMUST00000022053.11
|
Trip13
|
thyroid hormone receptor interactor 13 |
| chr18_-_60981981 | 14.90 |
ENSMUST00000177172.8
ENSMUST00000175934.8 ENSMUST00000176630.8 |
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chr5_-_138170644 | 14.67 |
ENSMUST00000000505.16
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr10_-_87982732 | 14.39 |
ENSMUST00000164121.8
ENSMUST00000164803.2 ENSMUST00000168163.8 ENSMUST00000048518.16 |
Parpbp
|
PARP1 binding protein |
| chr8_+_18645144 | 14.05 |
ENSMUST00000039412.15
|
Mcph1
|
microcephaly, primary autosomal recessive 1 |
| chr16_-_90524214 | 13.58 |
ENSMUST00000099554.5
|
Mis18a
|
MIS18 kinetochore protein A |
| chr11_-_101442663 | 13.39 |
ENSMUST00000017290.11
|
Brca1
|
breast cancer 1, early onset |
| chr1_-_189420270 | 13.37 |
ENSMUST00000171929.8
ENSMUST00000165962.2 |
Cenpf
|
centromere protein F |
| chr15_+_12824925 | 13.02 |
ENSMUST00000090292.13
|
Drosha
|
drosha, ribonuclease type III |
| chr12_+_105976463 | 12.81 |
ENSMUST00000072040.7
|
Vrk1
|
vaccinia related kinase 1 |
| chr15_+_12824901 | 12.76 |
ENSMUST00000169061.8
|
Drosha
|
drosha, ribonuclease type III |
| chr12_+_105976533 | 12.72 |
ENSMUST00000085026.12
ENSMUST00000021539.16 |
Vrk1
|
vaccinia related kinase 1 |
| chr2_+_129435448 | 12.62 |
ENSMUST00000049262.14
ENSMUST00000163034.8 ENSMUST00000160276.2 |
Sirpa
|
signal-regulatory protein alpha |
| chrX_-_165992145 | 12.61 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr3_-_54823287 | 12.56 |
ENSMUST00000070342.4
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr5_-_138170077 | 12.45 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chr6_-_56681657 | 12.05 |
ENSMUST00000176595.3
ENSMUST00000170382.5 ENSMUST00000203958.2 |
Lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr4_-_59783780 | 11.76 |
ENSMUST00000107526.8
ENSMUST00000095063.11 |
Inip
|
INTS3 and NABP interacting protein |
| chr12_+_117807607 | 11.71 |
ENSMUST00000176735.8
ENSMUST00000177339.2 |
Cdca7l
|
cell division cycle associated 7 like |
| chr7_+_126808016 | 11.20 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr2_+_180961507 | 11.04 |
ENSMUST00000098971.11
ENSMUST00000054622.15 ENSMUST00000108814.8 ENSMUST00000048608.16 ENSMUST00000108815.8 |
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr9_+_44045859 | 10.97 |
ENSMUST00000034650.15
ENSMUST00000098852.3 ENSMUST00000216002.2 |
Mcam
|
melanoma cell adhesion molecule |
| chr5_-_33809640 | 10.79 |
ENSMUST00000151081.2
ENSMUST00000139518.8 ENSMUST00000101354.10 |
Slbp
|
stem-loop binding protein |
| chr5_-_33809872 | 10.71 |
ENSMUST00000057551.14
|
Slbp
|
stem-loop binding protein |
| chr2_+_30176395 | 10.24 |
ENSMUST00000064447.12
|
Nup188
|
nucleoporin 188 |
| chr13_+_23719579 | 9.94 |
ENSMUST00000080859.8
|
H3c8
|
H3 clustered histone 8 |
| chr17_+_35219941 | 9.87 |
ENSMUST00000087315.14
|
Vars
|
valyl-tRNA synthetase |
| chrX_+_104123341 | 9.59 |
ENSMUST00000033577.11
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr3_-_10273628 | 9.46 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr17_+_35219998 | 9.35 |
ENSMUST00000173584.8
|
Vars
|
valyl-tRNA synthetase |
| chr2_+_129435115 | 9.34 |
ENSMUST00000099113.10
ENSMUST00000103202.10 |
Sirpa
|
signal-regulatory protein alpha |
| chr9_+_57615816 | 9.31 |
ENSMUST00000043990.14
ENSMUST00000142807.2 |
Edc3
|
enhancer of mRNA decapping 3 |
| chr1_+_9868332 | 9.21 |
ENSMUST00000166384.8
ENSMUST00000168907.8 |
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr5_-_33809683 | 9.13 |
ENSMUST00000075670.13
|
Slbp
|
stem-loop binding protein |
| chr2_-_3513783 | 9.09 |
ENSMUST00000124331.8
ENSMUST00000140494.2 ENSMUST00000027961.12 |
Gm45902
Hspa14
|
predicted gene 45902 heat shock protein 14 |
| chr12_+_99850756 | 9.01 |
ENSMUST00000153627.8
|
Tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr8_+_108271643 | 8.94 |
ENSMUST00000212543.2
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chrX_+_161684563 | 8.82 |
ENSMUST00000112303.8
ENSMUST00000033727.14 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr19_+_46064409 | 8.54 |
ENSMUST00000223728.2
ENSMUST00000235620.2 |
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chrX_+_161684221 | 8.44 |
ENSMUST00000101095.9
|
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr6_+_70703409 | 8.41 |
ENSMUST00000103410.3
|
Igkc
|
immunoglobulin kappa constant |
| chr14_+_76725876 | 8.36 |
ENSMUST00000101618.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr17_+_35220252 | 8.33 |
ENSMUST00000174260.8
|
Vars
|
valyl-tRNA synthetase |
| chr14_-_54754810 | 8.11 |
ENSMUST00000023873.12
|
Prmt5
|
protein arginine N-methyltransferase 5 |
| chr13_-_23929490 | 8.03 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3 |
| chr9_+_108820846 | 8.01 |
ENSMUST00000198140.5
ENSMUST00000051873.15 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr9_-_32255604 | 7.88 |
ENSMUST00000034533.7
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr2_+_24257576 | 7.69 |
ENSMUST00000140547.2
ENSMUST00000102942.8 |
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr4_-_3835595 | 7.54 |
ENSMUST00000138502.2
|
Rps20
|
ribosomal protein S20 |
| chr4_+_11558905 | 7.54 |
ENSMUST00000095145.12
ENSMUST00000108306.9 ENSMUST00000070755.13 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr15_-_98729333 | 7.20 |
ENSMUST00000168846.3
|
Prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr5_-_113910571 | 7.12 |
ENSMUST00000019118.8
|
Sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr7_-_126807581 | 7.10 |
ENSMUST00000120705.3
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr5_+_31046196 | 6.85 |
ENSMUST00000069705.11
ENSMUST00000201917.4 ENSMUST00000201168.4 ENSMUST00000202060.4 ENSMUST00000201225.4 ENSMUST00000114700.9 |
Agbl5
|
ATP/GTP binding protein-like 5 |
| chr6_-_36787096 | 6.78 |
ENSMUST00000201321.2
ENSMUST00000101534.5 |
Ptn
|
pleiotrophin |
| chr7_+_24920840 | 6.59 |
ENSMUST00000055604.6
|
Zfp526
|
zinc finger protein 526 |
| chr9_-_32255556 | 6.57 |
ENSMUST00000214223.2
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr4_-_43562397 | 6.53 |
ENSMUST00000030187.14
|
Tln1
|
talin 1 |
| chr5_-_25910788 | 6.39 |
ENSMUST00000030773.12
|
Xrcc2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chrX_+_161684735 | 6.38 |
ENSMUST00000112302.8
ENSMUST00000112301.8 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr11_-_106890307 | 6.24 |
ENSMUST00000018506.13
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr9_-_32255533 | 6.17 |
ENSMUST00000216033.2
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr17_+_35201074 | 6.00 |
ENSMUST00000007266.14
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr8_+_124750133 | 5.97 |
ENSMUST00000034457.9
|
Urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr3_-_142587419 | 5.96 |
ENSMUST00000174422.8
ENSMUST00000173830.8 |
Pkn2
|
protein kinase N2 |
| chr19_-_5416626 | 5.83 |
ENSMUST00000237167.2
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr2_+_129434834 | 5.76 |
ENSMUST00000103203.8
|
Sirpa
|
signal-regulatory protein alpha |
| chr8_+_18645543 | 5.71 |
ENSMUST00000146819.8
|
Mcph1
|
microcephaly, primary autosomal recessive 1 |
| chr8_-_106223502 | 5.47 |
ENSMUST00000212303.2
|
Zdhhc1
|
zinc finger, DHHC domain containing 1 |
| chr7_+_24584076 | 5.38 |
ENSMUST00000153451.9
ENSMUST00000108429.8 |
Rps19
|
ribosomal protein S19 |
| chr13_+_74085916 | 5.32 |
ENSMUST00000222399.2
ENSMUST00000099384.4 |
Brd9
|
bromodomain containing 9 |
| chr4_+_44004437 | 5.27 |
ENSMUST00000107846.10
|
Clta
|
clathrin, light polypeptide (Lca) |
| chr7_+_97020810 | 5.25 |
ENSMUST00000098300.6
|
Alg8
|
asparagine-linked glycosylation 8 (alpha-1,3-glucosyltransferase) |
| chr11_-_106890195 | 5.18 |
ENSMUST00000106768.2
ENSMUST00000144834.8 |
Kpna2
|
karyopherin (importin) alpha 2 |
| chr19_+_6451667 | 5.12 |
ENSMUST00000113471.3
ENSMUST00000113469.3 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr7_-_92523396 | 5.06 |
ENSMUST00000209074.2
ENSMUST00000208356.2 ENSMUST00000032877.11 |
Ddias
|
DNA damage-induced apoptosis suppressor |
| chrX_+_163763588 | 4.81 |
ENSMUST00000167446.8
ENSMUST00000057150.8 |
Fancb
|
Fanconi anemia, complementation group B |
| chr10_+_87982854 | 4.73 |
ENSMUST00000052355.15
|
Nup37
|
nucleoporin 37 |
| chr10_+_12936248 | 4.59 |
ENSMUST00000193426.6
|
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr10_+_36850532 | 4.58 |
ENSMUST00000019911.14
ENSMUST00000105510.2 |
Hdac2
|
histone deacetylase 2 |
| chr9_+_106354463 | 4.47 |
ENSMUST00000047721.10
|
Rrp9
|
ribosomal RNA processing 9, U3 small nucleolar RNA binding protein |
| chr1_+_52669849 | 4.43 |
ENSMUST00000165859.8
|
Nemp2
|
nuclear envelope integral membrane protein 2 |
| chr14_+_55815580 | 4.31 |
ENSMUST00000174484.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr17_+_35201130 | 4.30 |
ENSMUST00000173004.2
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_30176418 | 4.28 |
ENSMUST00000138666.8
ENSMUST00000113634.3 |
Nup188
|
nucleoporin 188 |
| chr10_+_87982916 | 4.00 |
ENSMUST00000169309.3
|
Nup37
|
nucleoporin 37 |
| chr12_-_4283926 | 3.94 |
ENSMUST00000111169.10
ENSMUST00000020981.12 |
Cenpo
|
centromere protein O |
| chr1_+_85822250 | 3.82 |
ENSMUST00000185569.2
|
Itm2c
|
integral membrane protein 2C |
| chr9_-_123507847 | 3.79 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr19_+_46064302 | 3.76 |
ENSMUST00000165017.2
ENSMUST00000223741.2 ENSMUST00000225780.2 |
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr17_+_25946644 | 3.44 |
ENSMUST00000237183.2
ENSMUST00000237785.2 ENSMUST00000047273.3 |
Rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr17_-_57338468 | 3.39 |
ENSMUST00000007814.10
ENSMUST00000233480.2 |
Khsrp
|
KH-type splicing regulatory protein |
| chr1_-_10108325 | 3.35 |
ENSMUST00000027050.10
ENSMUST00000188619.2 |
Cops5
|
COP9 signalosome subunit 5 |
| chr5_-_92496730 | 3.35 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr2_+_180961599 | 3.07 |
ENSMUST00000153112.2
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr2_+_71617266 | 3.06 |
ENSMUST00000112101.8
ENSMUST00000028522.10 |
Itga6
|
integrin alpha 6 |
| chr15_+_55171138 | 2.64 |
ENSMUST00000023053.12
ENSMUST00000110217.10 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr17_-_87332780 | 2.62 |
ENSMUST00000024957.7
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr1_+_92862432 | 2.16 |
ENSMUST00000117814.8
|
Capn10
|
calpain 10 |
| chr11_-_6556053 | 2.15 |
ENSMUST00000045713.4
|
Nacad
|
NAC alpha domain containing |
| chrX_+_104123367 | 2.14 |
ENSMUST00000119477.2
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr5_+_21577640 | 2.07 |
ENSMUST00000035799.6
|
Fgl2
|
fibrinogen-like protein 2 |
| chr2_+_106525938 | 1.96 |
ENSMUST00000016530.14
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr1_-_179631190 | 1.75 |
ENSMUST00000027768.14
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr19_+_8713156 | 1.68 |
ENSMUST00000210512.2
ENSMUST00000049424.11 |
Wdr74
|
WD repeat domain 74 |
| chr4_-_141553306 | 1.56 |
ENSMUST00000102481.4
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr2_-_145776934 | 1.52 |
ENSMUST00000001818.5
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chrX_+_12454031 | 1.50 |
ENSMUST00000033313.3
|
Atp6ap2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr1_-_74640504 | 1.46 |
ENSMUST00000136078.2
ENSMUST00000132081.2 ENSMUST00000113721.8 ENSMUST00000027357.12 |
Rnf25
|
ring finger protein 25 |
| chr2_+_125994050 | 1.26 |
ENSMUST00000170908.8
|
Dtwd1
|
DTW domain containing 1 |
| chr17_-_84495364 | 1.17 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr2_+_112114906 | 1.16 |
ENSMUST00000053666.8
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr15_+_57775595 | 1.12 |
ENSMUST00000022992.13
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr13_+_74086241 | 0.97 |
ENSMUST00000222749.2
|
Brd9
|
bromodomain containing 9 |
| chr3_-_116456292 | 0.96 |
ENSMUST00000029570.9
|
Mfsd14a
|
major facilitator superfamily domain containing 14A |
| chr5_+_31046670 | 0.92 |
ENSMUST00000200850.2
|
Agbl5
|
ATP/GTP binding protein-like 5 |
| chr9_-_82857528 | 0.88 |
ENSMUST00000034787.12
|
Phip
|
pleckstrin homology domain interacting protein |
| chr14_+_30741082 | 0.86 |
ENSMUST00000112098.11
ENSMUST00000112095.8 ENSMUST00000112106.8 ENSMUST00000146325.8 |
Pbrm1
|
polybromo 1 |
| chr7_-_16019379 | 0.78 |
ENSMUST00000174270.8
|
Ccdc9
|
coiled-coil domain containing 9 |
| chr1_-_77491683 | 0.75 |
ENSMUST00000186930.2
ENSMUST00000027451.13 ENSMUST00000188797.7 |
Epha4
|
Eph receptor A4 |
| chr17_+_87332978 | 0.42 |
ENSMUST00000024959.10
|
Cript
|
cysteine-rich PDZ-binding protein |
| chr12_-_118162652 | 0.25 |
ENSMUST00000084806.7
|
Dnah11
|
dynein, axonemal, heavy chain 11 |
| chr17_+_35200823 | 0.19 |
ENSMUST00000114011.11
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_-_126859790 | 0.10 |
ENSMUST00000035276.5
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr10_+_17598961 | 0.09 |
ENSMUST00000038107.9
ENSMUST00000219558.2 ENSMUST00000218370.2 |
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr19_+_5416769 | 0.08 |
ENSMUST00000025759.9
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr3_-_142587678 | 0.05 |
ENSMUST00000043812.15
|
Pkn2
|
protein kinase N2 |
| chr6_-_24956296 | 0.05 |
ENSMUST00000127247.4
|
Tmem229a
|
transmembrane protein 229A |
| chr17_+_31515163 | 0.01 |
ENSMUST00000235972.2
ENSMUST00000165149.3 ENSMUST00000236251.2 |
Slc37a1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.1 | 39.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 6.4 | 25.5 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 6.1 | 30.7 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 5.2 | 20.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 4.6 | 27.6 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 4.6 | 18.2 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 3.9 | 69.9 | GO:0015816 | glycine transport(GO:0015816) |
| 3.5 | 14.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 3.2 | 25.8 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 3.1 | 99.9 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 2.8 | 19.8 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 2.8 | 50.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 2.8 | 30.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 2.7 | 8.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 2.7 | 13.4 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 2.5 | 22.1 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 2.3 | 13.6 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 2.3 | 27.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 2.3 | 6.8 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) positive regulation of neuromuscular junction development(GO:1904398) |
| 2.1 | 14.5 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 2.1 | 20.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 1.9 | 19.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 1.9 | 18.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.8 | 73.9 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 1.7 | 15.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 1.6 | 33.9 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 1.6 | 23.6 | GO:0006241 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 1.4 | 17.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 1.4 | 14.9 | GO:0014029 | neural crest formation(GO:0014029) |
| 1.4 | 17.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 1.3 | 36.0 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 1.2 | 41.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 1.2 | 5.8 | GO:0015074 | DNA integration(GO:0015074) |
| 1.2 | 17.3 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 1.0 | 27.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 1.0 | 9.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.9 | 41.4 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.9 | 4.6 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.8 | 3.3 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.8 | 30.6 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.8 | 8.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.8 | 12.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.8 | 7.8 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.7 | 9.0 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.5 | 9.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.5 | 24.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.5 | 3.8 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.5 | 11.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.5 | 6.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.5 | 5.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.4 | 13.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.4 | 28.8 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.4 | 13.4 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.4 | 2.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.3 | 3.4 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) tRNA pseudouridine synthesis(GO:0031119) |
| 0.3 | 27.7 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.3 | 3.4 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.3 | 3.9 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.3 | 1.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.3 | 35.2 | GO:0007051 | spindle organization(GO:0007051) |
| 0.3 | 1.2 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.3 | 4.6 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.3 | 5.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.2 | 0.7 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.2 | 6.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 1.5 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.2 | 7.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 2.2 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 0.9 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.2 | 9.2 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 5.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 5.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 7.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 4.3 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.1 | 11.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 11.2 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.1 | 0.9 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 7.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 11.8 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.1 | 0.4 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 8.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.6 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 2.6 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.1 | 9.3 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 41.5 | GO:0051301 | cell division(GO:0051301) |
| 0.1 | 5.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 2.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 2.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.3 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 5.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 1.7 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 17.1 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 1.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 11.4 | GO:0030036 | actin cytoskeleton organization(GO:0030036) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 25.8 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 7.5 | 45.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 6.6 | 39.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 5.0 | 29.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 4.8 | 28.8 | GO:0031523 | Myb complex(GO:0031523) |
| 3.8 | 30.6 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 3.4 | 13.5 | GO:0035101 | FACT complex(GO:0035101) |
| 3.0 | 20.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 2.6 | 18.2 | GO:0005638 | lamin filament(GO:0005638) |
| 2.6 | 30.7 | GO:0008278 | cohesin complex(GO:0008278) |
| 2.5 | 22.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 2.4 | 7.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 2.2 | 22.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 2.1 | 27.1 | GO:0042555 | MCM complex(GO:0042555) |
| 2.1 | 14.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 2.0 | 11.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 1.8 | 5.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 1.2 | 13.4 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 1.0 | 13.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.8 | 10.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.8 | 15.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.7 | 4.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.7 | 2.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.7 | 8.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.6 | 41.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.5 | 4.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 4.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.4 | 50.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.3 | 15.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.3 | 4.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 12.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 35.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.2 | 7.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 56.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.2 | 25.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 1.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 10.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 20.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 27.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 1.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 11.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 7.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 6.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 11.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 14.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 6.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 12.2 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 44.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 8.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 9.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 7.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 6.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 3.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 15.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 6.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 8.3 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 50.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 12.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 24.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 4.4 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 111.3 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 1.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 25.5 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 7.7 | 30.6 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 6.9 | 20.8 | GO:0003896 | DNA primase activity(GO:0003896) |
| 6.3 | 44.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 5.9 | 23.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 5.7 | 33.9 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 5.2 | 25.8 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 5.0 | 29.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 5.0 | 14.9 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 4.7 | 66.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 4.6 | 27.6 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 3.5 | 24.4 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 2.9 | 20.6 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 2.4 | 19.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 2.4 | 28.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 2.3 | 39.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 2.0 | 22.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 1.7 | 5.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.7 | 6.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.6 | 8.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 1.5 | 4.6 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 1.1 | 41.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 1.1 | 7.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.9 | 41.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.9 | 7.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.9 | 8.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.6 | 3.8 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.6 | 18.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.6 | 18.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.6 | 3.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.5 | 30.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.5 | 4.5 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.5 | 14.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.4 | 6.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 7.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.4 | 4.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 9.3 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.4 | 11.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.4 | 10.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.4 | 13.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.4 | 9.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.4 | 5.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 30.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 1.2 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.3 | 6.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.3 | 12.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 7.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 27.7 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.2 | 2.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 13.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 3.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 7.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 5.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 11.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 5.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 23.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.2 | 1.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 3.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 8.6 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 2.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 7.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 8.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 9.7 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 3.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 7.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 6.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 4.7 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 3.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 5.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 15.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 38.6 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 41.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 1.5 | 62.6 | PID ATM PATHWAY | ATM pathway |
| 1.1 | 51.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.8 | 66.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.7 | 27.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 20.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.4 | 13.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.4 | 11.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 44.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 6.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 18.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 12.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 34.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.2 | 5.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 4.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 30.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 3.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 7.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 5.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 4.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 71.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 2.6 | 30.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 2.4 | 30.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 2.3 | 36.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 2.1 | 20.8 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 2.1 | 18.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.9 | 27.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.5 | 28.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 1.4 | 31.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 1.2 | 18.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 1.0 | 66.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 1.0 | 25.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.9 | 27.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.9 | 98.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.8 | 9.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.8 | 27.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.8 | 5.8 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.8 | 23.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.6 | 58.6 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.5 | 7.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.5 | 6.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.4 | 8.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.4 | 15.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.4 | 20.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.4 | 9.0 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.3 | 9.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 5.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 12.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 8.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 5.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 3.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 12.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 6.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 4.3 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 3.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 4.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |