Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Gata2_Gata1
Z-value: 5.28
Transcription factors associated with Gata2_Gata1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
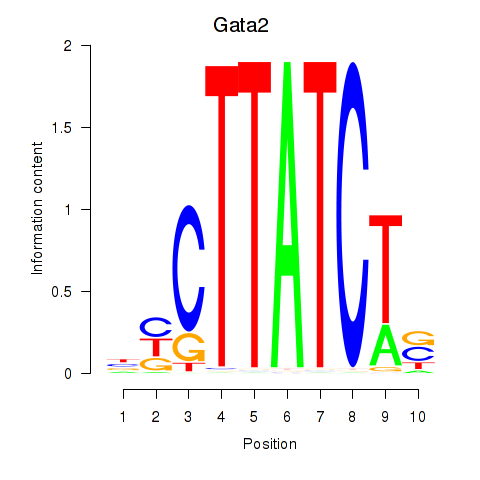
Gata2
|
ENSMUSG00000015053.15 | GATA binding protein 2 |
|
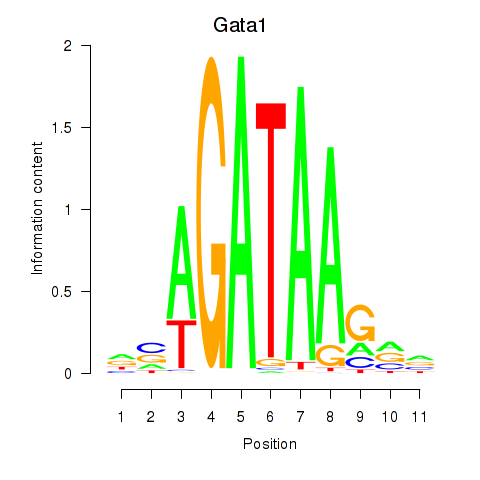
Gata1
|
ENSMUSG00000031162.15 | GATA binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata1 | mm39_v1_chrX_-_7834057_7834149 | 0.87 | 9.3e-12 | Click! |
| Gata2 | mm39_v1_chr6_+_88175312_88175316 | 0.81 | 2.7e-09 | Click! |
Activity profile of Gata2_Gata1 motif
Sorted Z-values of Gata2_Gata1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_85428059 | 95.47 |
ENSMUST00000238364.2
ENSMUST00000238562.2 ENSMUST00000037165.6 |
Lyl1
|
lymphoblastomic leukemia 1 |
| chr17_-_26417982 | 88.11 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr6_+_30639217 | 87.16 |
ENSMUST00000031806.10
|
Cpa1
|
carboxypeptidase A1, pancreatic |
| chr17_-_28779678 | 87.07 |
ENSMUST00000114785.3
ENSMUST00000025062.5 |
Clps
|
colipase, pancreatic |
| chr2_+_84810802 | 86.84 |
ENSMUST00000028467.6
|
Prg2
|
proteoglycan 2, bone marrow |
| chr1_-_132295617 | 82.84 |
ENSMUST00000142609.8
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr17_+_37180437 | 81.60 |
ENSMUST00000060524.11
|
Trim10
|
tripartite motif-containing 10 |
| chr4_-_46404224 | 75.96 |
ENSMUST00000107764.9
|
Hemgn
|
hemogen |
| chr9_+_20940669 | 74.58 |
ENSMUST00000001040.7
ENSMUST00000215077.2 |
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr6_+_41369290 | 72.20 |
ENSMUST00000049079.9
|
Gm5771
|
predicted gene 5771 |
| chr15_-_103159892 | 71.17 |
ENSMUST00000133600.8
ENSMUST00000134554.2 ENSMUST00000156927.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr6_+_41435846 | 69.62 |
ENSMUST00000031910.8
|
Prss1
|
protease, serine 1 (trypsin 1) |
| chr6_+_41279199 | 67.40 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr2_-_120867529 | 67.10 |
ENSMUST00000102490.10
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr4_-_137157824 | 65.91 |
ENSMUST00000102522.5
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr15_-_103160082 | 65.35 |
ENSMUST00000149111.8
ENSMUST00000132836.8 |
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr9_-_44253588 | 63.97 |
ENSMUST00000215091.2
|
Hmbs
|
hydroxymethylbilane synthase |
| chr6_-_41681273 | 63.50 |
ENSMUST00000031899.14
|
Kel
|
Kell blood group |
| chr10_-_62178453 | 62.21 |
ENSMUST00000143179.2
ENSMUST00000130422.8 |
Hk1
|
hexokinase 1 |
| chr9_-_21874802 | 59.36 |
ENSMUST00000006397.7
|
Epor
|
erythropoietin receptor |
| chr6_+_30541581 | 59.10 |
ENSMUST00000096066.5
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr4_+_134591847 | 58.47 |
ENSMUST00000030627.8
|
Rhd
|
Rh blood group, D antigen |
| chr11_-_102360664 | 53.94 |
ENSMUST00000103086.4
|
Itga2b
|
integrin alpha 2b |
| chr2_-_120867232 | 52.89 |
ENSMUST00000023987.6
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr2_+_103788321 | 48.84 |
ENSMUST00000156813.8
ENSMUST00000170926.8 |
Lmo2
|
LIM domain only 2 |
| chr11_+_95227836 | 47.57 |
ENSMUST00000037502.7
|
Fam117a
|
family with sequence similarity 117, member A |
| chr7_-_98887770 | 43.99 |
ENSMUST00000064231.8
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr17_-_35304582 | 43.87 |
ENSMUST00000038507.7
|
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr6_+_41331039 | 43.20 |
ENSMUST00000072103.7
|
Try10
|
trypsin 10 |
| chr8_+_85428391 | 40.67 |
ENSMUST00000238338.2
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr9_-_44253630 | 39.84 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr19_-_6996791 | 39.28 |
ENSMUST00000040772.9
|
Fermt3
|
fermitin family member 3 |
| chrX_-_106446928 | 38.48 |
ENSMUST00000033591.6
|
Itm2a
|
integral membrane protein 2A |
| chr11_-_83177548 | 36.43 |
ENSMUST00000163961.3
|
Slfn14
|
schlafen 14 |
| chr6_-_41291634 | 35.94 |
ENSMUST00000064324.12
|
Try5
|
trypsin 5 |
| chr6_+_41498716 | 35.02 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr6_+_86055018 | 34.54 |
ENSMUST00000205034.3
ENSMUST00000203724.3 |
Add2
|
adducin 2 (beta) |
| chr16_+_38182569 | 34.48 |
ENSMUST00000023494.13
ENSMUST00000114739.2 |
Popdc2
|
popeye domain containing 2 |
| chr7_+_99184645 | 33.99 |
ENSMUST00000098266.9
ENSMUST00000179755.8 |
Arrb1
|
arrestin, beta 1 |
| chr11_+_32226400 | 33.93 |
ENSMUST00000020531.9
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr19_-_6996696 | 33.76 |
ENSMUST00000236188.2
|
Fermt3
|
fermitin family member 3 |
| chrX_+_149330371 | 31.32 |
ENSMUST00000066337.13
ENSMUST00000112715.2 |
Alas2
|
aminolevulinic acid synthase 2, erythroid |
| chr8_-_86107593 | 28.81 |
ENSMUST00000122452.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr6_+_86055048 | 27.44 |
ENSMUST00000032069.8
|
Add2
|
adducin 2 (beta) |
| chr2_-_153083322 | 27.31 |
ENSMUST00000056924.14
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr14_-_70945434 | 27.08 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr17_-_33932346 | 25.93 |
ENSMUST00000173392.2
|
Marchf2
|
membrane associated ring-CH-type finger 2 |
| chrX_-_7830405 | 23.68 |
ENSMUST00000128449.2
|
Gata1
|
GATA binding protein 1 |
| chr3_-_107129038 | 23.42 |
ENSMUST00000029504.9
|
Cym
|
chymosin |
| chr7_+_28240262 | 22.74 |
ENSMUST00000119180.4
|
Sycn
|
syncollin |
| chr11_+_32226893 | 21.84 |
ENSMUST00000145569.2
|
Hba-x
|
hemoglobin X, alpha-like embryonic chain in Hba complex |
| chr4_-_119047202 | 21.53 |
ENSMUST00000239029.2
ENSMUST00000138395.9 ENSMUST00000156746.3 |
Ermap
|
erythroblast membrane-associated protein |
| chr18_-_78166595 | 21.00 |
ENSMUST00000091813.12
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr11_+_4936824 | 19.81 |
ENSMUST00000109897.8
ENSMUST00000009234.16 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr14_+_26722319 | 19.58 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr11_-_99328969 | 19.26 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chr18_-_78166539 | 18.88 |
ENSMUST00000160292.8
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr2_+_72306503 | 18.82 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr3_-_20296337 | 18.55 |
ENSMUST00000001921.3
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr19_-_6065181 | 18.37 |
ENSMUST00000236537.2
ENSMUST00000025891.11 |
Capn1
|
calpain 1 |
| chr5_+_115604321 | 18.14 |
ENSMUST00000145785.8
ENSMUST00000031495.11 ENSMUST00000112071.8 ENSMUST00000125568.2 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr9_-_57743989 | 17.41 |
ENSMUST00000164010.8
ENSMUST00000171444.8 ENSMUST00000098686.4 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chrX_+_92718695 | 17.41 |
ENSMUST00000045898.4
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr4_-_119047167 | 17.40 |
ENSMUST00000030396.15
|
Ermap
|
erythroblast membrane-associated protein |
| chr17_-_31363245 | 17.15 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr19_-_6065415 | 16.64 |
ENSMUST00000237519.2
|
Capn1
|
calpain 1 |
| chr11_-_115968745 | 16.31 |
ENSMUST00000156545.2
ENSMUST00000075036.9 ENSMUST00000106451.8 |
Unc13d
|
unc-13 homolog D |
| chr15_+_57849269 | 16.05 |
ENSMUST00000050374.3
|
Fam83a
|
family with sequence similarity 83, member A |
| chr11_+_70529944 | 15.99 |
ENSMUST00000055184.7
ENSMUST00000108551.3 |
Gp1ba
|
glycoprotein 1b, alpha polypeptide |
| chr4_-_137137088 | 15.69 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr17_-_35285146 | 15.47 |
ENSMUST00000174190.2
ENSMUST00000097337.8 |
Mpig6b
|
megakaryocyte and platelet inhibitory receptor G6b |
| chr10_-_62163444 | 15.41 |
ENSMUST00000139228.8
|
Hk1
|
hexokinase 1 |
| chr4_-_87724533 | 15.24 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr11_+_70396070 | 14.77 |
ENSMUST00000019063.3
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr6_-_136899167 | 14.65 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr10_+_79855454 | 14.49 |
ENSMUST00000043311.7
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr11_-_115968373 | 14.28 |
ENSMUST00000174822.8
|
Unc13d
|
unc-13 homolog D |
| chr8_-_86091946 | 14.14 |
ENSMUST00000034133.14
|
Mylk3
|
myosin light chain kinase 3 |
| chr3_+_108272205 | 13.93 |
ENSMUST00000090563.7
|
Mybphl
|
myosin binding protein H-like |
| chr8_-_86091970 | 13.48 |
ENSMUST00000121972.8
|
Mylk3
|
myosin light chain kinase 3 |
| chr4_-_119047146 | 13.45 |
ENSMUST00000124626.9
|
Ermap
|
erythroblast membrane-associated protein |
| chr4_-_119047180 | 13.43 |
ENSMUST00000150864.3
ENSMUST00000141227.9 |
Ermap
|
erythroblast membrane-associated protein |
| chr6_-_136918885 | 13.42 |
ENSMUST00000111891.4
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr1_-_173161069 | 13.40 |
ENSMUST00000038227.6
|
Ackr1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr16_+_58490625 | 12.78 |
ENSMUST00000060077.7
|
Cpox
|
coproporphyrinogen oxidase |
| chr6_-_136918495 | 12.74 |
ENSMUST00000111892.8
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr4_-_59438633 | 12.67 |
ENSMUST00000040166.14
ENSMUST00000107544.2 |
Susd1
|
sushi domain containing 1 |
| chr5_-_73349191 | 12.66 |
ENSMUST00000176910.3
|
Fryl
|
FRY like transcription coactivator |
| chr6_+_87405968 | 12.43 |
ENSMUST00000032125.7
|
Bmp10
|
bone morphogenetic protein 10 |
| chr6_-_136918844 | 11.95 |
ENSMUST00000204934.2
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr10_-_80257681 | 11.95 |
ENSMUST00000156244.2
|
Tcf3
|
transcription factor 3 |
| chr9_+_66257747 | 11.78 |
ENSMUST00000042824.13
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr6_-_136918671 | 11.65 |
ENSMUST00000032344.12
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr11_-_115968576 | 11.57 |
ENSMUST00000106450.8
|
Unc13d
|
unc-13 homolog D |
| chr3_-_14873406 | 10.52 |
ENSMUST00000181860.8
ENSMUST00000144327.3 |
Car1
|
carbonic anhydrase 1 |
| chr15_+_80507671 | 10.04 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr11_-_115967873 | 9.94 |
ENSMUST00000153408.8
|
Unc13d
|
unc-13 homolog D |
| chr2_+_84867554 | 9.51 |
ENSMUST00000077798.13
|
Ssrp1
|
structure specific recognition protein 1 |
| chr7_+_110372860 | 9.41 |
ENSMUST00000143786.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr10_-_80269436 | 9.15 |
ENSMUST00000105346.10
ENSMUST00000020377.13 ENSMUST00000105340.8 ENSMUST00000020379.13 ENSMUST00000105344.8 ENSMUST00000105342.8 ENSMUST00000105345.10 ENSMUST00000105343.8 |
Tcf3
|
transcription factor 3 |
| chr8_+_121215155 | 9.02 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr6_+_125529911 | 8.65 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr7_-_119058489 | 8.52 |
ENSMUST00000207887.3
ENSMUST00000239424.2 ENSMUST00000033255.8 |
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr15_-_98505508 | 8.50 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr4_-_87724512 | 8.42 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr19_-_6065799 | 8.34 |
ENSMUST00000235138.2
|
Capn1
|
calpain 1 |
| chr15_+_76788270 | 8.12 |
ENSMUST00000004072.10
ENSMUST00000229183.2 |
Rpl8
|
ribosomal protein L8 |
| chr2_+_84867783 | 8.06 |
ENSMUST00000168266.8
ENSMUST00000130729.3 |
Ssrp1
|
structure specific recognition protein 1 |
| chr1_-_138103021 | 7.97 |
ENSMUST00000182755.8
ENSMUST00000193650.2 ENSMUST00000182283.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr7_-_4400704 | 7.77 |
ENSMUST00000108590.4
ENSMUST00000206928.2 |
Gp6
|
glycoprotein 6 (platelet) |
| chr1_-_45965661 | 7.69 |
ENSMUST00000186804.2
ENSMUST00000187406.7 ENSMUST00000187420.7 |
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr2_-_156848923 | 7.40 |
ENSMUST00000146413.8
ENSMUST00000103129.9 ENSMUST00000103130.8 |
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr7_+_110226857 | 7.35 |
ENSMUST00000033054.10
|
Adm
|
adrenomedullin |
| chr11_+_4959515 | 7.26 |
ENSMUST00000101613.3
|
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr8_+_71047110 | 7.16 |
ENSMUST00000019283.10
ENSMUST00000210005.2 |
Isyna1
|
myo-inositol 1-phosphate synthase A1 |
| chr2_-_73316053 | 6.99 |
ENSMUST00000102680.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr15_+_55171138 | 5.87 |
ENSMUST00000023053.12
ENSMUST00000110217.10 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr8_+_121264161 | 5.74 |
ENSMUST00000118136.2
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr10_+_39009951 | 5.70 |
ENSMUST00000019991.8
|
Tube1
|
tubulin, epsilon 1 |
| chr14_-_54891073 | 5.56 |
ENSMUST00000126166.8
ENSMUST00000141453.8 ENSMUST00000150371.8 ENSMUST00000123875.2 ENSMUST00000022794.14 ENSMUST00000148754.10 |
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr1_-_138102972 | 5.28 |
ENSMUST00000195533.6
ENSMUST00000183301.8 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr4_+_40920047 | 5.04 |
ENSMUST00000030122.5
|
Spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr5_+_32616187 | 5.03 |
ENSMUST00000015100.15
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr15_+_102392586 | 4.97 |
ENSMUST00000229061.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr9_+_48073296 | 4.93 |
ENSMUST00000216998.2
ENSMUST00000215780.2 |
Nxpe4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr5_+_64969679 | 4.82 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr16_+_44914397 | 4.73 |
ENSMUST00000061050.6
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr9_-_44624496 | 4.71 |
ENSMUST00000144251.8
ENSMUST00000156918.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr1_-_160134873 | 4.34 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr11_-_94133527 | 4.27 |
ENSMUST00000061469.4
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr10_+_5543769 | 4.20 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr12_-_69939931 | 4.09 |
ENSMUST00000049239.8
ENSMUST00000110570.8 |
Map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr9_-_65734826 | 4.01 |
ENSMUST00000159109.2
|
Zfp609
|
zinc finger protein 609 |
| chr3_-_75177378 | 3.96 |
ENSMUST00000039047.5
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr11_-_75060345 | 3.65 |
ENSMUST00000055619.5
|
Hic1
|
hypermethylated in cancer 1 |
| chr17_+_71511642 | 3.63 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr16_+_44913974 | 3.57 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr13_-_73848807 | 3.56 |
ENSMUST00000022048.6
|
Slc6a19
|
solute carrier family 6 (neurotransmitter transporter), member 19 |
| chr17_-_82045800 | 3.44 |
ENSMUST00000235015.2
ENSMUST00000163123.3 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr15_-_38518458 | 3.38 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr9_+_56982622 | 3.36 |
ENSMUST00000167715.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr19_-_60770628 | 3.33 |
ENSMUST00000238125.2
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr1_+_40468720 | 3.31 |
ENSMUST00000174335.8
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr3_+_89979948 | 3.29 |
ENSMUST00000121503.8
ENSMUST00000119570.8 |
Tpm3
|
tropomyosin 3, gamma |
| chr3_+_89043879 | 3.24 |
ENSMUST00000107482.10
ENSMUST00000127058.2 |
Pklr
|
pyruvate kinase liver and red blood cell |
| chr3_+_89043440 | 3.21 |
ENSMUST00000047111.13
|
Pklr
|
pyruvate kinase liver and red blood cell |
| chr1_+_45350698 | 3.05 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr11_-_86248395 | 2.95 |
ENSMUST00000043624.9
|
Med13
|
mediator complex subunit 13 |
| chr16_-_63684477 | 2.84 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chr10_+_58207229 | 2.80 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr14_-_79718890 | 2.69 |
ENSMUST00000022601.7
|
Wbp4
|
WW domain binding protein 4 |
| chr7_-_44541787 | 2.48 |
ENSMUST00000208551.2
ENSMUST00000208253.2 ENSMUST00000207654.2 ENSMUST00000207278.2 |
Med25
|
mediator complex subunit 25 |
| chr1_+_75483721 | 2.48 |
ENSMUST00000037330.5
|
Inha
|
inhibin alpha |
| chr8_-_65489834 | 2.45 |
ENSMUST00000142822.4
|
Apela
|
apelin receptor early endogenous ligand |
| chr1_+_12762501 | 2.43 |
ENSMUST00000177608.8
ENSMUST00000180062.8 |
Sulf1
|
sulfatase 1 |
| chr15_-_77037756 | 2.35 |
ENSMUST00000227314.2
ENSMUST00000227930.2 ENSMUST00000227533.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr7_-_44542098 | 2.34 |
ENSMUST00000003049.8
|
Med25
|
mediator complex subunit 25 |
| chrX_-_104919201 | 2.33 |
ENSMUST00000198209.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr7_-_141241632 | 2.30 |
ENSMUST00000239500.1
|
ENSMUSG00000118661.1
|
mucin 6, gastric |
| chr4_-_132649798 | 2.29 |
ENSMUST00000097856.10
ENSMUST00000030696.11 |
Fam76a
|
family with sequence similarity 76, member A |
| chr5_+_24569802 | 2.29 |
ENSMUST00000115090.6
ENSMUST00000030834.7 |
Nos3
|
nitric oxide synthase 3, endothelial cell |
| chr15_-_77037972 | 2.14 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr2_+_112097087 | 2.12 |
ENSMUST00000110987.9
ENSMUST00000028549.14 |
Slc12a6
|
solute carrier family 12, member 6 |
| chr5_-_122402451 | 2.11 |
ENSMUST00000111738.8
|
Tctn1
|
tectonic family member 1 |
| chr14_-_54647647 | 2.07 |
ENSMUST00000228488.2
ENSMUST00000195970.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr11_-_53598078 | 1.76 |
ENSMUST00000124352.2
ENSMUST00000020649.14 |
Rad50
|
RAD50 double strand break repair protein |
| chr2_+_112114906 | 1.72 |
ENSMUST00000053666.8
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr5_-_35683035 | 1.70 |
ENSMUST00000038676.7
|
Cpz
|
carboxypeptidase Z |
| chr8_+_85635189 | 1.64 |
ENSMUST00000003910.13
ENSMUST00000109744.8 |
Dnase2a
|
deoxyribonuclease II alpha |
| chr8_-_65489791 | 1.60 |
ENSMUST00000124790.8
|
Apela
|
apelin receptor early endogenous ligand |
| chr19_-_37153436 | 1.58 |
ENSMUST00000142973.2
ENSMUST00000154376.8 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr9_+_109881083 | 1.57 |
ENSMUST00000164930.8
ENSMUST00000199498.5 |
Map4
|
microtubule-associated protein 4 |
| chr5_-_125371162 | 1.56 |
ENSMUST00000127148.2
|
Scarb1
|
scavenger receptor class B, member 1 |
| chr8_+_85635341 | 1.48 |
ENSMUST00000134569.2
|
Dnase2a
|
deoxyribonuclease II alpha |
| chr6_+_17306334 | 1.45 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr11_-_5848771 | 1.44 |
ENSMUST00000102921.4
|
Myl7
|
myosin, light polypeptide 7, regulatory |
| chr9_-_32452885 | 1.42 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr1_-_106980033 | 1.30 |
ENSMUST00000112717.3
|
Serpinb3a
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A |
| chr2_-_86109346 | 1.26 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr2_+_109522781 | 1.23 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr4_+_124696336 | 1.19 |
ENSMUST00000138807.8
ENSMUST00000030723.3 |
Mtf1
|
metal response element binding transcription factor 1 |
| chr3_+_105811712 | 1.15 |
ENSMUST00000000574.3
|
Adora3
|
adenosine A3 receptor |
| chr16_-_95260104 | 1.00 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr18_+_76192529 | 0.95 |
ENSMUST00000167921.2
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr10_-_13264574 | 0.92 |
ENSMUST00000079698.7
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr8_+_71047010 | 0.90 |
ENSMUST00000211117.2
|
Isyna1
|
myo-inositol 1-phosphate synthase A1 |
| chr15_-_82796308 | 0.89 |
ENSMUST00000109510.10
ENSMUST00000048966.7 |
Tcf20
|
transcription factor 20 |
| chr3_-_102690026 | 0.87 |
ENSMUST00000172026.8
ENSMUST00000170856.6 |
Tshb
|
thyroid stimulating hormone, beta subunit |
| chr2_-_20973692 | 0.86 |
ENSMUST00000114594.8
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr17_-_84495364 | 0.80 |
ENSMUST00000060366.7
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr18_+_57275854 | 0.77 |
ENSMUST00000139892.2
|
Megf10
|
multiple EGF-like-domains 10 |
| chr2_+_3705824 | 0.77 |
ENSMUST00000115054.9
|
Fam107b
|
family with sequence similarity 107, member B |
| chr8_-_40095706 | 0.76 |
ENSMUST00000210525.2
ENSMUST00000170091.3 |
Msr1
|
macrophage scavenger receptor 1 |
| chr16_-_63684425 | 0.75 |
ENSMUST00000232049.2
|
Epha3
|
Eph receptor A3 |
| chr1_+_172168764 | 0.72 |
ENSMUST00000056136.4
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr8_+_71047263 | 0.72 |
ENSMUST00000211501.2
|
Isyna1
|
myo-inositol 1-phosphate synthase A1 |
| chr8_+_85635311 | 0.69 |
ENSMUST00000145292.8
|
Dnase2a
|
deoxyribonuclease II alpha |
| chr10_+_90412827 | 0.69 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr8_+_11608412 | 0.60 |
ENSMUST00000209565.2
|
Ing1
|
inhibitor of growth family, member 1 |
| chr8_-_40095661 | 0.58 |
ENSMUST00000026021.14
|
Msr1
|
macrophage scavenger receptor 1 |
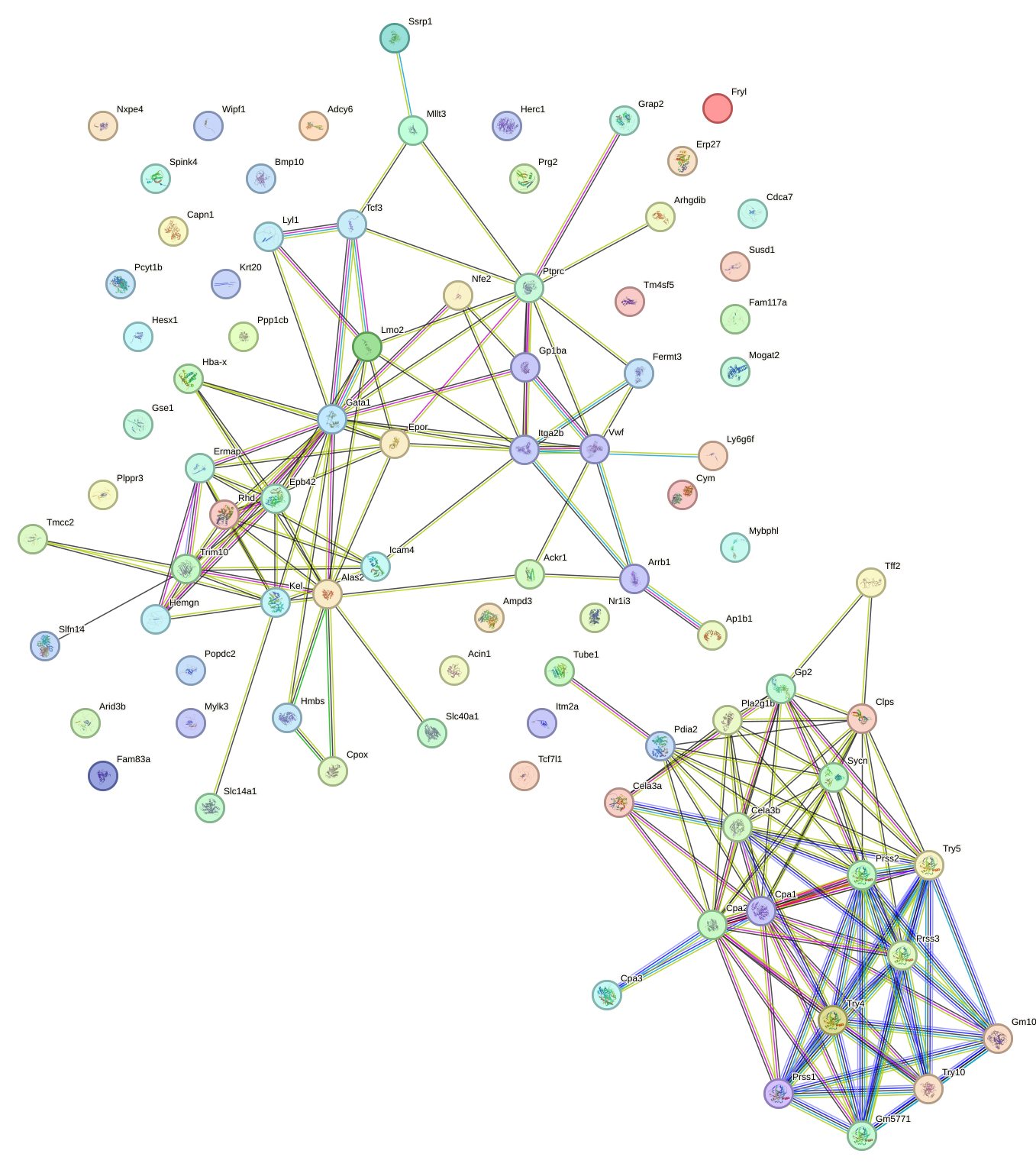
Network of associatons between targets according to the STRING database.
First level regulatory network of Gata2_Gata1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 34.6 | 103.8 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 28.9 | 86.8 | GO:0002215 | defense response to nematode(GO:0002215) |
| 14.7 | 44.0 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 14.6 | 58.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 11.3 | 136.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 10.6 | 63.5 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 9.7 | 77.6 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 9.4 | 56.4 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 9.3 | 175.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 9.1 | 27.3 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 9.1 | 136.5 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 7.9 | 23.7 | GO:0030221 | basophil differentiation(GO:0030221) |
| 7.3 | 73.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 6.9 | 34.5 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 6.5 | 52.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 6.2 | 37.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 6.0 | 18.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 5.5 | 49.8 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 5.5 | 38.5 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 4.2 | 34.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 4.2 | 59.4 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 3.5 | 48.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 3.3 | 83.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 3.3 | 13.3 | GO:2000473 | regulation of hematopoietic stem cell migration(GO:2000471) positive regulation of hematopoietic stem cell migration(GO:2000473) |
| 2.9 | 8.8 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 2.8 | 8.5 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 2.8 | 8.5 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 2.6 | 21.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 2.6 | 33.8 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 2.6 | 7.7 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 2.6 | 12.8 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 2.4 | 17.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 2.3 | 43.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 1.9 | 7.4 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 1.8 | 77.2 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 1.8 | 19.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 1.8 | 84.3 | GO:0032094 | response to food(GO:0032094) |
| 1.7 | 38.5 | GO:0031000 | response to caffeine(GO:0031000) |
| 1.7 | 36.4 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 1.7 | 62.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 1.6 | 4.8 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 1.4 | 18.5 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 1.2 | 7.5 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 1.2 | 3.6 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 1.1 | 82.8 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 1.1 | 23.7 | GO:0007379 | segment specification(GO:0007379) |
| 1.0 | 3.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.0 | 3.8 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.9 | 9.4 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.9 | 7.4 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.9 | 27.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.9 | 12.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.8 | 127.7 | GO:0007586 | digestion(GO:0007586) |
| 0.8 | 3.4 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.8 | 3.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.8 | 3.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.8 | 5.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.8 | 2.3 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.7 | 11.8 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.7 | 64.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.6 | 6.4 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.5 | 1.6 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.5 | 5.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.5 | 2.9 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.4 | 2.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.4 | 7.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.4 | 1.2 | GO:0061193 | taste bud development(GO:0061193) |
| 0.4 | 5.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.4 | 34.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.4 | 0.7 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.4 | 2.8 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 | 1.3 | GO:0010885 | regulation of cholesterol storage(GO:0010885) positive regulation of cholesterol storage(GO:0010886) |
| 0.3 | 0.3 | GO:0048807 | female genitalia morphogenesis(GO:0048807) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.3 | 4.7 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.3 | 3.3 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.3 | 40.4 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.3 | 5.0 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.3 | 5.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.3 | 3.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.3 | 0.8 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 1.5 | GO:0035127 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.2 | 13.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 0.5 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.2 | 1.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 32.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.2 | 1.0 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 | 8.6 | GO:0030168 | platelet activation(GO:0030168) |
| 0.2 | 10.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 3.0 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.2 | 0.9 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 1.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.2 | 2.9 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 11.6 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 8.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 1.1 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 2.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 5.9 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 16.1 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 0.8 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 0.5 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 3.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 4.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 8.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.3 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.1 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 21.1 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 72.8 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 3.8 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 3.6 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.3 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.0 | GO:0009750 | response to fructose(GO:0009750) |
| 0.0 | 4.3 | GO:0006260 | DNA replication(GO:0006260) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 55.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 5.6 | 55.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 4.1 | 62.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 2.5 | 7.4 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 1.8 | 77.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 1.8 | 21.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 1.6 | 33.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.2 | 70.0 | GO:0002102 | podosome(GO:0002102) |
| 1.1 | 3.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 1.1 | 54.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.8 | 2.5 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 0.7 | 128.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.6 | 7.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.6 | 120.0 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.6 | 5.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.6 | 16.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.6 | 5.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.6 | 27.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.4 | 1.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.4 | 23.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.4 | 105.3 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.3 | 8.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 27.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.3 | 566.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 62.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 3.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 12.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 17.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 3.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 41.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 0.7 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 27.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 8.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 31.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 48.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 8.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 7.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 2.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 10.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 53.3 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.1 | 5.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 11.5 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 132.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 22.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 5.7 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 34.6 | 103.8 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 12.0 | 120.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 11.3 | 34.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 10.8 | 53.9 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 10.4 | 31.3 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 9.7 | 77.6 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 7.3 | 88.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 7.3 | 44.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 6.3 | 56.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 6.2 | 55.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 5.7 | 39.9 | GO:0015265 | urea channel activity(GO:0015265) |
| 5.5 | 49.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 5.3 | 165.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 4.4 | 17.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 2.9 | 8.8 | GO:0004512 | inositol-3-phosphate synthase activity(GO:0004512) intramolecular lyase activity(GO:0016872) |
| 2.6 | 21.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 2.3 | 134.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 2.1 | 27.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 2.0 | 18.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 1.9 | 58.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 1.7 | 367.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 1.4 | 43.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 1.3 | 23.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 1.3 | 6.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.2 | 17.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 1.2 | 8.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.1 | 3.4 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 1.1 | 12.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 1.1 | 3.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 1.1 | 3.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 1.1 | 12.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.1 | 10.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.0 | 3.8 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 1.0 | 13.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.9 | 9.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.8 | 3.8 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.7 | 34.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.7 | 60.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.6 | 14.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.6 | 107.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.6 | 45.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.6 | 7.8 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.6 | 2.3 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.5 | 8.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.5 | 56.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.5 | 16.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.5 | 38.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.4 | 13.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.4 | 1.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.4 | 2.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 29.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.4 | 3.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.4 | 5.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.3 | 43.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.3 | 8.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 2.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.3 | 0.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 4.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 56.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.2 | 27.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.2 | 1.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 4.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 61.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.2 | 1.8 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.2 | 3.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 10.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 68.4 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.1 | 8.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.8 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.5 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 13.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 10.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 3.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 3.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 4.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 61.0 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.1 | 3.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 5.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 8.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 7.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 33.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 2.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 9.7 | GO:0003682 | chromatin binding(GO:0003682) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 74.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.9 | 86.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 1.9 | 54.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.8 | 102.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.7 | 34.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.7 | 10.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.7 | 178.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.7 | 41.6 | PID EPO PATHWAY | EPO signaling pathway |
| 0.6 | 62.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.6 | 37.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.6 | 21.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.5 | 49.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.4 | 8.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 25.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.3 | 13.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.3 | 24.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 3.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 2.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 3.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 4.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 8.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 11.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 5.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 6.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 147.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 3.0 | 93.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 2.8 | 62.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 1.8 | 27.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 1.4 | 34.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 1.3 | 13.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 1.3 | 77.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 1.2 | 16.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.2 | 68.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 1.0 | 18.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.9 | 33.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.8 | 138.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.8 | 21.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.5 | 17.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.5 | 8.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.4 | 9.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.4 | 3.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.4 | 3.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 10.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 16.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 44.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 7.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 5.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 6.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 5.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 7.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 11.4 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.1 | 5.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 5.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |