Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
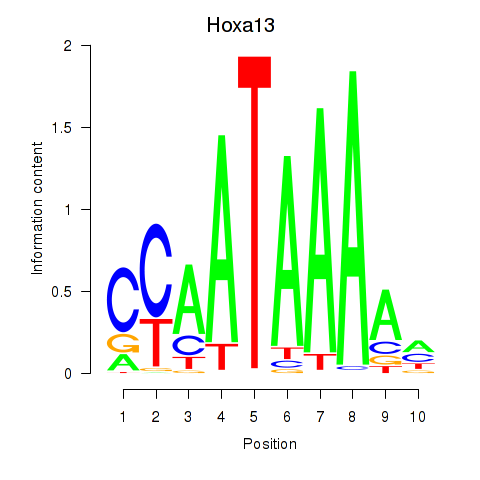
Results for Hoxa13
Z-value: 2.31
Transcription factors associated with Hoxa13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxa13
|
ENSMUSG00000038203.22 | homeobox A13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxa13 | mm39_v1_chr6_-_52237765_52237788 | -0.02 | 8.9e-01 | Click! |
Activity profile of Hoxa13 motif
Sorted Z-values of Hoxa13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_61592331 | 44.93 |
ENSMUST00000098040.4
|
Mup18
|
major urinary protein 18 |
| chr4_-_61259997 | 43.63 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chr4_-_61259801 | 40.41 |
ENSMUST00000125461.8
|
Mup14
|
major urinary protein 14 |
| chr19_+_40078132 | 34.10 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr4_-_60070411 | 33.17 |
ENSMUST00000079697.10
ENSMUST00000125282.2 ENSMUST00000166098.8 |
Mup7
|
major urinary protein 7 |
| chr4_-_60538151 | 32.22 |
ENSMUST00000098047.3
|
Mup10
|
major urinary protein 10 |
| chr4_-_61437704 | 31.63 |
ENSMUST00000095051.6
ENSMUST00000107483.8 |
Mup16
|
major urinary protein 16 |
| chr4_-_60377932 | 31.40 |
ENSMUST00000107506.9
ENSMUST00000122381.8 ENSMUST00000118759.8 ENSMUST00000132829.3 |
Mup9
|
major urinary protein 9 |
| chr4_-_60618357 | 31.34 |
ENSMUST00000084544.5
ENSMUST00000098046.10 |
Mup11
|
major urinary protein 11 |
| chr4_-_61184270 | 31.24 |
ENSMUST00000072678.6
ENSMUST00000098042.10 |
Mup13
|
major urinary protein 13 |
| chr4_-_61357980 | 30.67 |
ENSMUST00000095049.5
|
Mup15
|
major urinary protein 15 |
| chr7_-_30623592 | 29.92 |
ENSMUST00000217812.2
ENSMUST00000074671.9 |
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr4_-_60777462 | 29.85 |
ENSMUST00000211875.2
|
Mup22
|
major urinary protein 22 |
| chr4_-_60139857 | 29.20 |
ENSMUST00000107490.5
ENSMUST00000074700.9 |
Mup2
|
major urinary protein 2 |
| chr4_-_60222580 | 27.45 |
ENSMUST00000095058.5
ENSMUST00000163931.8 |
Mup8
|
major urinary protein 8 |
| chr4_-_61514108 | 27.02 |
ENSMUST00000107484.2
|
Mup17
|
major urinary protein 17 |
| chr8_-_25066313 | 25.64 |
ENSMUST00000121992.2
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr4_-_60697274 | 25.15 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr4_-_60457902 | 24.22 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr15_+_6474808 | 21.37 |
ENSMUST00000022749.17
ENSMUST00000239466.2 |
C9
|
complement component 9 |
| chr8_-_94006345 | 18.43 |
ENSMUST00000034178.9
|
Ces1f
|
carboxylesterase 1F |
| chr5_-_87485023 | 16.73 |
ENSMUST00000031195.3
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr12_-_72675624 | 14.24 |
ENSMUST00000208039.2
ENSMUST00000207585.2 |
Gm4756
|
predicted gene 4756 |
| chr19_-_8109346 | 12.76 |
ENSMUST00000065651.5
|
Slc22a28
|
solute carrier family 22, member 28 |
| chr19_+_12610870 | 11.16 |
ENSMUST00000119960.2
|
Glyat
|
glycine-N-acyltransferase |
| chr19_+_12610668 | 10.19 |
ENSMUST00000044976.12
|
Glyat
|
glycine-N-acyltransferase |
| chr7_-_19432129 | 10.00 |
ENSMUST00000172808.2
ENSMUST00000174191.2 |
Apoe
|
apolipoprotein E |
| chr13_-_24098981 | 9.37 |
ENSMUST00000110407.4
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr2_-_86180622 | 9.01 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr14_+_66207163 | 8.98 |
ENSMUST00000153460.8
|
Clu
|
clusterin |
| chr13_-_24098951 | 8.46 |
ENSMUST00000021769.16
|
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr5_-_89583469 | 8.41 |
ENSMUST00000200534.2
|
Gc
|
vitamin D binding protein |
| chr10_-_115198093 | 8.17 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr1_-_121260298 | 7.80 |
ENSMUST00000071064.13
|
Insig2
|
insulin induced gene 2 |
| chr1_-_192946359 | 7.65 |
ENSMUST00000161737.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chrX_+_59044796 | 7.60 |
ENSMUST00000033477.5
|
F9
|
coagulation factor IX |
| chr7_-_30643444 | 7.46 |
ENSMUST00000062620.9
|
Hamp
|
hepcidin antimicrobial peptide |
| chr19_+_38995463 | 7.21 |
ENSMUST00000025966.5
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr5_-_145946408 | 6.60 |
ENSMUST00000138870.2
ENSMUST00000068317.13 |
Cyp3a25
|
cytochrome P450, family 3, subfamily a, polypeptide 25 |
| chr14_+_66205932 | 6.60 |
ENSMUST00000022616.14
|
Clu
|
clusterin |
| chr10_-_115197775 | 6.45 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr1_-_121260274 | 6.39 |
ENSMUST00000161068.2
|
Insig2
|
insulin induced gene 2 |
| chr10_-_108846816 | 6.21 |
ENSMUST00000105276.8
ENSMUST00000064054.14 |
Syt1
|
synaptotagmin I |
| chr14_-_47426863 | 6.16 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr11_-_11848107 | 6.14 |
ENSMUST00000178704.8
|
Ddc
|
dopa decarboxylase |
| chr10_-_116732813 | 6.05 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr19_-_6117815 | 5.99 |
ENSMUST00000162575.8
ENSMUST00000159084.8 ENSMUST00000161718.8 ENSMUST00000162810.8 ENSMUST00000025713.12 ENSMUST00000113543.9 ENSMUST00000160417.8 ENSMUST00000161528.2 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr17_+_31652029 | 5.87 |
ENSMUST00000136384.9
|
Pde9a
|
phosphodiesterase 9A |
| chr1_-_72251466 | 5.82 |
ENSMUST00000048860.9
|
Mreg
|
melanoregulin |
| chr17_+_31652073 | 5.81 |
ENSMUST00000237363.2
|
Pde9a
|
phosphodiesterase 9A |
| chr11_-_73215442 | 5.78 |
ENSMUST00000021119.9
|
Aspa
|
aspartoacylase |
| chr5_+_146016064 | 5.73 |
ENSMUST00000035571.10
|
Cyp3a59
|
cytochrome P450, family 3, subfamily a, polypeptide 59 |
| chr19_+_24853039 | 5.54 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr12_-_72675670 | 5.39 |
ENSMUST00000209038.2
|
Gm4756
|
predicted gene 4756 |
| chr1_+_167135933 | 5.37 |
ENSMUST00000195015.6
|
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr14_-_61794330 | 5.34 |
ENSMUST00000022497.15
|
Spryd7
|
SPRY domain containing 7 |
| chr15_+_3300249 | 5.32 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr18_+_38551960 | 5.29 |
ENSMUST00000236085.2
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr8_-_5155347 | 5.21 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr15_+_54975713 | 5.12 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chrM_+_3906 | 5.01 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr8_+_96078886 | 4.83 |
ENSMUST00000034243.7
|
Mmp15
|
matrix metallopeptidase 15 |
| chr14_-_52150804 | 4.61 |
ENSMUST00000004673.15
ENSMUST00000111632.5 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr2_+_162829250 | 4.60 |
ENSMUST00000018012.14
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr16_-_23807602 | 4.59 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr18_+_38552011 | 4.53 |
ENSMUST00000025293.5
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr2_+_162829422 | 4.48 |
ENSMUST00000117123.2
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr3_-_146321341 | 4.45 |
ENSMUST00000200633.2
|
Dnase2b
|
deoxyribonuclease II beta |
| chr4_+_148225139 | 4.37 |
ENSMUST00000140049.8
ENSMUST00000105707.2 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr5_-_87686048 | 4.30 |
ENSMUST00000031199.11
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr7_+_127399776 | 4.18 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr4_+_41762309 | 4.12 |
ENSMUST00000108042.3
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr2_-_91025441 | 4.11 |
ENSMUST00000002177.9
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr11_-_113600838 | 4.03 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr9_-_44710480 | 3.94 |
ENSMUST00000214833.2
ENSMUST00000213972.2 ENSMUST00000214431.2 ENSMUST00000213363.2 ENSMUST00000114705.9 ENSMUST00000002100.8 |
Tmem25
|
transmembrane protein 25 |
| chr3_-_10396418 | 3.92 |
ENSMUST00000191670.6
ENSMUST00000065938.15 ENSMUST00000118410.8 |
Impa1
|
inositol (myo)-1(or 4)-monophosphatase 1 |
| chr12_-_86773160 | 3.91 |
ENSMUST00000021682.9
|
Angel1
|
angel homolog 1 |
| chr10_+_116137277 | 3.87 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr1_+_175459735 | 3.72 |
ENSMUST00000097458.4
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_-_91025380 | 3.67 |
ENSMUST00000111356.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr1_+_175459559 | 3.64 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr1_+_185064339 | 3.60 |
ENSMUST00000027916.13
ENSMUST00000210277.2 ENSMUST00000151769.2 ENSMUST00000110965.2 |
Bpnt1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr12_+_59176543 | 3.57 |
ENSMUST00000069430.15
ENSMUST00000177370.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr19_+_39499288 | 3.53 |
ENSMUST00000025968.5
|
Cyp2c39
|
cytochrome P450, family 2, subfamily c, polypeptide 39 |
| chr7_-_30555592 | 3.52 |
ENSMUST00000185748.2
ENSMUST00000094583.2 |
Ffar3
|
free fatty acid receptor 3 |
| chr17_-_36207965 | 3.50 |
ENSMUST00000150056.2
ENSMUST00000156817.2 ENSMUST00000146451.8 ENSMUST00000148482.8 |
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr19_+_26725589 | 3.42 |
ENSMUST00000207812.2
ENSMUST00000175791.9 ENSMUST00000207118.2 ENSMUST00000209085.2 ENSMUST00000112637.10 ENSMUST00000207054.2 ENSMUST00000208589.2 ENSMUST00000176475.9 ENSMUST00000176698.9 ENSMUST00000207832.2 ENSMUST00000177252.9 ENSMUST00000208712.2 ENSMUST00000208186.2 ENSMUST00000208806.2 ENSMUST00000208027.2 |
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_+_45960804 | 3.41 |
ENSMUST00000067065.14
ENSMUST00000124544.8 ENSMUST00000138049.9 ENSMUST00000132139.9 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_+_129157576 | 3.34 |
ENSMUST00000032260.6
|
Clec2d
|
C-type lectin domain family 2, member d |
| chr2_-_91025208 | 3.28 |
ENSMUST00000111355.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr13_+_23868175 | 3.22 |
ENSMUST00000018246.6
|
H2bc4
|
H2B clustered histone 4 |
| chr8_-_25528972 | 3.18 |
ENSMUST00000084031.6
|
Htra4
|
HtrA serine peptidase 4 |
| chr2_-_91025492 | 3.12 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr10_-_12689345 | 3.09 |
ENSMUST00000217899.2
|
Utrn
|
utrophin |
| chr4_+_98919183 | 3.08 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr19_+_46695889 | 3.06 |
ENSMUST00000003655.9
|
As3mt
|
arsenite methyltransferase |
| chr10_-_68114543 | 3.03 |
ENSMUST00000219238.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr7_+_119206233 | 3.00 |
ENSMUST00000126367.8
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr11_+_96920751 | 2.99 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr8_+_45960931 | 2.99 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr3_-_89319088 | 2.98 |
ENSMUST00000107429.10
ENSMUST00000129308.9 ENSMUST00000107426.8 ENSMUST00000050398.11 ENSMUST00000162701.2 |
Flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr12_-_104120105 | 2.94 |
ENSMUST00000085050.4
|
Serpina3c
|
serine (or cysteine) peptidase inhibitor, clade A, member 3C |
| chr12_+_59176506 | 2.91 |
ENSMUST00000175912.8
ENSMUST00000176892.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr2_-_144392696 | 2.89 |
ENSMUST00000028915.6
|
Rbbp9
|
retinoblastoma binding protein 9, serine hydrolase |
| chr8_+_4399588 | 2.87 |
ENSMUST00000110982.8
ENSMUST00000024004.9 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr11_+_96085118 | 2.84 |
ENSMUST00000062709.4
|
Hoxb13
|
homeobox B13 |
| chrX_+_60753074 | 2.84 |
ENSMUST00000075983.6
|
Ldoc1
|
regulator of NFKB signaling |
| chr18_-_3281752 | 2.83 |
ENSMUST00000140332.8
ENSMUST00000147138.8 |
Crem
|
cAMP responsive element modulator |
| chr13_-_24118139 | 2.82 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr11_-_17161504 | 2.79 |
ENSMUST00000020317.8
|
Pno1
|
partner of NOB1 homolog |
| chr7_+_80707328 | 2.78 |
ENSMUST00000107348.2
|
Alpk3
|
alpha-kinase 3 |
| chr4_-_108075119 | 2.75 |
ENSMUST00000223127.2
ENSMUST00000043793.7 ENSMUST00000106690.9 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr11_-_120618052 | 2.74 |
ENSMUST00000106148.10
ENSMUST00000026144.5 |
Dcxr
|
dicarbonyl L-xylulose reductase |
| chr7_-_28079678 | 2.67 |
ENSMUST00000051241.7
|
Zfp36
|
zinc finger protein 36 |
| chrX_-_50889955 | 2.66 |
ENSMUST00000069509.4
ENSMUST00000114869.8 |
Usp26
|
ubiquitin specific peptidase 26 |
| chr1_-_36283326 | 2.65 |
ENSMUST00000046875.14
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr8_-_110305672 | 2.60 |
ENSMUST00000074898.8
|
Hp
|
haptoglobin |
| chr5_-_28672091 | 2.59 |
ENSMUST00000002708.5
|
Shh
|
sonic hedgehog |
| chr6_-_124942366 | 2.58 |
ENSMUST00000129446.8
ENSMUST00000032220.15 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr15_-_74869684 | 2.57 |
ENSMUST00000190188.2
ENSMUST00000189068.7 ENSMUST00000186526.7 ENSMUST00000187171.2 ENSMUST00000187994.7 |
Ly6a
|
lymphocyte antigen 6 complex, locus A |
| chr6_+_79794899 | 2.54 |
ENSMUST00000179797.3
|
Gm20594
|
predicted gene, 20594 |
| chr12_-_72132168 | 2.53 |
ENSMUST00000019862.3
|
L3hypdh
|
L-3-hydroxyproline dehydratase (trans-) |
| chr13_+_32985990 | 2.53 |
ENSMUST00000021832.7
|
Wrnip1
|
Werner helicase interacting protein 1 |
| chr13_-_14237958 | 2.51 |
ENSMUST00000223174.2
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr9_-_123461593 | 2.50 |
ENSMUST00000026273.11
|
Slc6a20b
|
solute carrier family 6 (neurotransmitter transporter), member 20B |
| chr5_+_150119860 | 2.48 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein |
| chrX_+_138464065 | 2.47 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr12_-_103623354 | 2.45 |
ENSMUST00000152517.2
|
Serpina6
|
serine (or cysteine) peptidase inhibitor, clade A, member 6 |
| chr13_-_64460491 | 2.45 |
ENSMUST00000222570.2
ENSMUST00000220895.2 |
Prxl2c
|
peroxiredoxin like 2C |
| chr3_+_62327089 | 2.45 |
ENSMUST00000161057.2
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr2_-_18053595 | 2.42 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr2_-_38816229 | 2.39 |
ENSMUST00000076275.11
ENSMUST00000142130.2 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr9_-_51240201 | 2.36 |
ENSMUST00000039959.11
ENSMUST00000238450.3 |
1810046K07Rik
|
RIKEN cDNA 1810046K07 gene |
| chr16_+_29398165 | 2.35 |
ENSMUST00000161186.8
ENSMUST00000038867.13 |
Opa1
|
OPA1, mitochondrial dynamin like GTPase |
| chr5_-_120610828 | 2.33 |
ENSMUST00000052258.14
ENSMUST00000031594.13 |
Sdsl
|
serine dehydratase-like |
| chrM_-_14061 | 2.33 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr6_+_82018604 | 2.31 |
ENSMUST00000042974.15
|
Eva1a
|
eva-1 homolog A (C. elegans) |
| chrX_+_149981074 | 2.25 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr17_-_36208265 | 2.22 |
ENSMUST00000148721.8
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr5_-_25910758 | 2.19 |
ENSMUST00000134972.3
|
Xrcc2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr7_-_73187369 | 2.11 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr4_+_89055359 | 2.07 |
ENSMUST00000058030.10
|
Mtap
|
methylthioadenosine phosphorylase |
| chr1_-_57008986 | 2.06 |
ENSMUST00000176759.2
ENSMUST00000177424.2 |
Satb2
|
special AT-rich sequence binding protein 2 |
| chr5_-_25047577 | 2.03 |
ENSMUST00000030787.9
|
Rheb
|
Ras homolog enriched in brain |
| chr17_+_53786240 | 2.03 |
ENSMUST00000017975.7
|
Rab5a
|
RAB5A, member RAS oncogene family |
| chr8_-_41507808 | 2.02 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr13_-_43634695 | 2.01 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr2_+_89804937 | 2.00 |
ENSMUST00000214630.2
ENSMUST00000111512.10 ENSMUST00000144710.3 ENSMUST00000216678.2 |
Olfr1260
|
olfactory receptor 1260 |
| chr2_-_63014622 | 1.98 |
ENSMUST00000075052.10
ENSMUST00000112454.8 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr16_+_41353360 | 1.98 |
ENSMUST00000099761.10
|
Lsamp
|
limbic system-associated membrane protein |
| chr3_+_20039775 | 1.96 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr6_+_145561483 | 1.95 |
ENSMUST00000087445.7
|
Tuba3b
|
tubulin, alpha 3B |
| chrX_+_37689503 | 1.95 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr2_-_91067212 | 1.94 |
ENSMUST00000111352.8
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr7_+_105290203 | 1.93 |
ENSMUST00000210893.2
|
Gm45799
|
predicted gene 45799 |
| chr12_+_44268134 | 1.92 |
ENSMUST00000122902.8
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr9_+_107217786 | 1.92 |
ENSMUST00000042581.4
|
6430571L13Rik
|
RIKEN cDNA 6430571L13 gene |
| chr16_+_34470290 | 1.91 |
ENSMUST00000148562.8
|
Ropn1
|
ropporin, rhophilin associated protein 1 |
| chr15_-_101336669 | 1.91 |
ENSMUST00000081945.5
|
Krt87
|
keratin 87 |
| chr9_+_45230370 | 1.90 |
ENSMUST00000034597.8
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr13_+_93810911 | 1.87 |
ENSMUST00000048001.8
|
Dmgdh
|
dimethylglycine dehydrogenase precursor |
| chr1_-_78488795 | 1.87 |
ENSMUST00000170511.3
|
BC035947
|
cDNA sequence BC035947 |
| chr2_-_69172944 | 1.87 |
ENSMUST00000102709.8
ENSMUST00000102710.10 ENSMUST00000180142.2 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr9_+_107765320 | 1.86 |
ENSMUST00000191906.6
ENSMUST00000035202.4 |
Mon1a
|
MON1 homolog A, secretory traffciking associated |
| chr16_+_91022300 | 1.84 |
ENSMUST00000035608.10
|
Olig2
|
oligodendrocyte transcription factor 2 |
| chr11_-_54751738 | 1.83 |
ENSMUST00000144164.9
|
Lyrm7
|
LYR motif containing 7 |
| chr13_+_19374502 | 1.82 |
ENSMUST00000198330.2
ENSMUST00000103555.3 |
Trgv6
|
T cell receptor gamma, variable 6 |
| chr6_-_52217821 | 1.81 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr10_-_116808514 | 1.80 |
ENSMUST00000092165.5
|
Gm10271
|
predicted gene 10271 |
| chr7_+_43079512 | 1.78 |
ENSMUST00000004732.7
|
Lim2
|
lens intrinsic membrane protein 2 |
| chr3_-_154036180 | 1.77 |
ENSMUST00000177846.8
|
Lhx8
|
LIM homeobox protein 8 |
| chr19_-_11058452 | 1.75 |
ENSMUST00000025636.8
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr13_-_64460382 | 1.74 |
ENSMUST00000021938.11
ENSMUST00000221118.2 ENSMUST00000221350.2 |
Prxl2c
|
peroxiredoxin like 2C |
| chr2_-_52566583 | 1.74 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr1_-_63215952 | 1.73 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr17_+_26048010 | 1.73 |
ENSMUST00000026832.14
|
Jmjd8
|
jumonji domain containing 8 |
| chr3_+_137377285 | 1.73 |
ENSMUST00000166899.3
|
Gm21962
|
predicted gene, 21962 |
| chr2_-_63014514 | 1.71 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr14_-_68771138 | 1.68 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr16_+_56024676 | 1.68 |
ENSMUST00000160116.8
ENSMUST00000069936.8 |
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr6_+_147377326 | 1.67 |
ENSMUST00000203659.3
ENSMUST00000032441.14 |
Ccdc91
|
coiled-coil domain containing 91 |
| chr7_-_135130374 | 1.67 |
ENSMUST00000053716.8
|
Clrn3
|
clarin 3 |
| chr17_-_26063488 | 1.67 |
ENSMUST00000176709.2
|
Rhot2
|
ras homolog family member T2 |
| chr3_+_96127174 | 1.66 |
ENSMUST00000073115.5
|
H2ac21
|
H2A clustered histone 21 |
| chr15_+_101371353 | 1.65 |
ENSMUST00000088049.5
|
Krt86
|
keratin 86 |
| chr4_-_81360993 | 1.65 |
ENSMUST00000107262.8
ENSMUST00000102830.10 |
Mpdz
|
multiple PDZ domain crumbs cell polarity complex component |
| chr3_+_138019040 | 1.62 |
ENSMUST00000013455.13
ENSMUST00000106247.2 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr13_+_34918820 | 1.62 |
ENSMUST00000039605.8
|
Fam50b
|
family with sequence similarity 50, member B |
| chr2_-_84508385 | 1.61 |
ENSMUST00000189772.2
ENSMUST00000053664.9 ENSMUST00000111664.8 |
Gm28635
Tmx2
|
predicted gene 28635 thioredoxin-related transmembrane protein 2 |
| chr11_+_98337655 | 1.59 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr6_-_124942170 | 1.58 |
ENSMUST00000148485.2
ENSMUST00000129976.8 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr3_-_96104886 | 1.58 |
ENSMUST00000016087.4
|
Bola1
|
bolA-like 1 (E. coli) |
| chr16_-_89305201 | 1.58 |
ENSMUST00000056118.4
|
Krtap7-1
|
keratin associated protein 7-1 |
| chr7_-_7281231 | 1.57 |
ENSMUST00000209833.2
ENSMUST00000209325.2 |
Gm45844
|
predicted gene 45844 |
| chr13_+_25127127 | 1.56 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr13_+_104246259 | 1.56 |
ENSMUST00000160322.8
ENSMUST00000159574.2 |
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr3_+_3699205 | 1.55 |
ENSMUST00000108394.3
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr7_+_43339842 | 1.55 |
ENSMUST00000056329.7
|
Klk14
|
kallikrein related-peptidase 14 |
| chr6_-_113577606 | 1.55 |
ENSMUST00000035870.5
|
Fancd2os
|
Fancd2 opposite strand |
| chr15_-_76116245 | 1.52 |
ENSMUST00000167754.8
|
Plec
|
plectin |
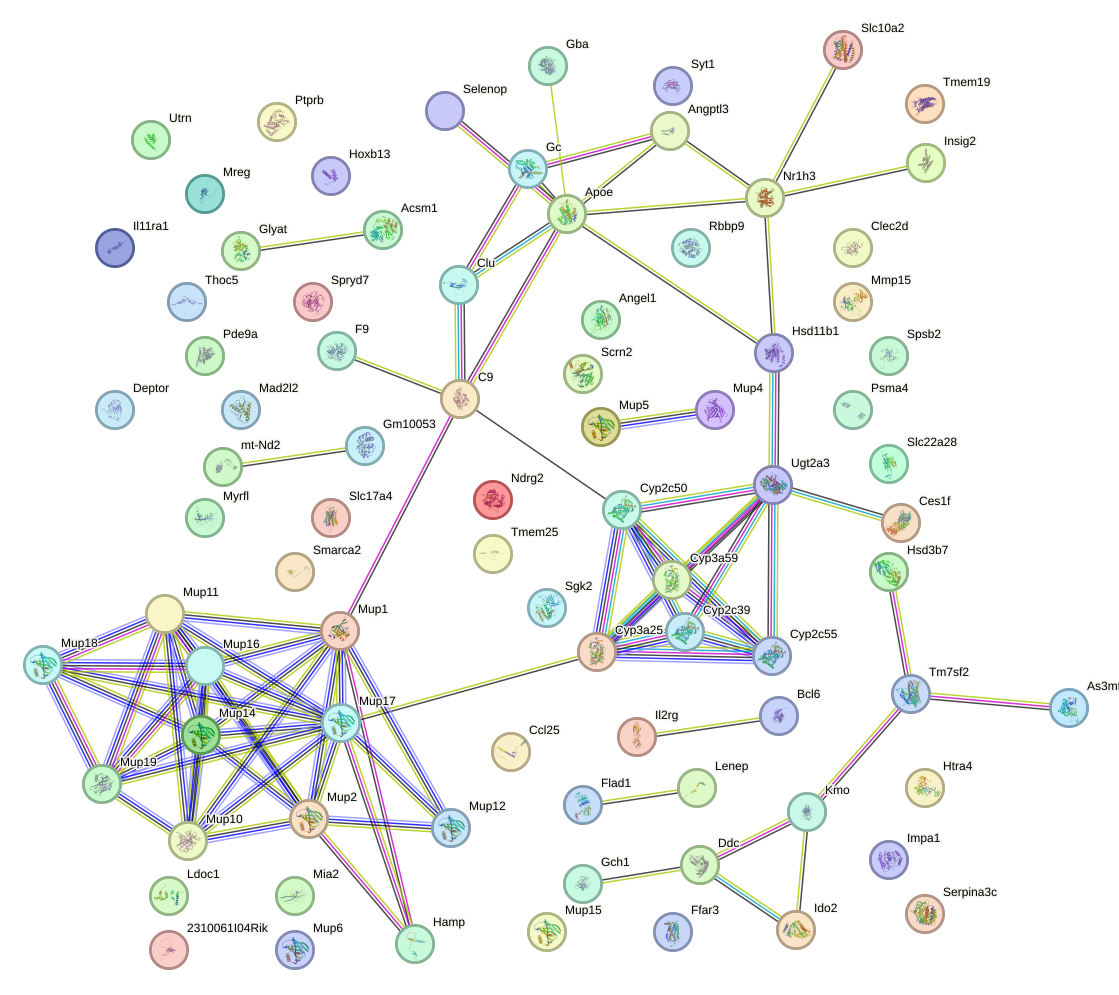
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 37.4 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 6.1 | 18.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 4.6 | 55.6 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 3.6 | 43.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 3.5 | 14.2 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 3.3 | 10.0 | GO:1903000 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 3.0 | 33.0 | GO:0042436 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
| 2.6 | 15.6 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 2.5 | 7.6 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 2.4 | 14.4 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 1.5 | 6.2 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 1.4 | 4.3 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 1.4 | 1.4 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 1.3 | 21.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.0 | 16.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 1.0 | 3.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.9 | 2.7 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.9 | 2.7 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.9 | 3.5 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.9 | 6.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.9 | 4.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.9 | 2.6 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.9 | 2.6 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) kidney smooth muscle tissue development(GO:0072194) negative regulation of dopaminergic neuron differentiation(GO:1904339) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.9 | 12.9 | GO:0015747 | urate transport(GO:0015747) |
| 0.8 | 15.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.8 | 5.8 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.8 | 4.6 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.8 | 2.3 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.7 | 2.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.7 | 2.8 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.7 | 6.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.7 | 5.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.6 | 1.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.6 | 2.5 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.6 | 1.8 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.6 | 1.7 | GO:0097037 | heme export(GO:0097037) |
| 0.6 | 12.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.5 | 4.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.5 | 1.6 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.5 | 3.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.5 | 1.5 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.5 | 1.5 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.5 | 1.4 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.5 | 1.0 | GO:0003130 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.5 | 2.3 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.5 | 1.4 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.4 | 3.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.4 | 3.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.4 | 5.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.4 | 1.2 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.4 | 1.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.4 | 1.2 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.4 | 1.2 | GO:0097115 | cytoskeletal matrix organization at active zone(GO:0048789) neurexin clustering involved in presynaptic membrane assembly(GO:0097115) retrograde trans-synaptic signaling by trans-synaptic protein complex(GO:0098942) |
| 0.4 | 1.1 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.4 | 4.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.4 | 1.9 | GO:0046618 | drug export(GO:0046618) |
| 0.4 | 4.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.4 | 0.7 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.4 | 4.0 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.4 | 0.7 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.4 | 2.5 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.4 | 7.5 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.4 | 1.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.3 | 7.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 1.3 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.3 | 1.0 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.3 | 1.9 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 3.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 5.7 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.3 | 2.8 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.3 | 1.9 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.3 | 5.8 | GO:0042640 | anagen(GO:0042640) |
| 0.3 | 2.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.3 | 2.0 | GO:2000286 | regulation of endosome size(GO:0051036) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 | 5.9 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 0.7 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 2.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 0.7 | GO:0048372 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.2 | 1.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 1.5 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.2 | 4.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 1.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 0.6 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.2 | 1.7 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.2 | 2.0 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.2 | 0.9 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.2 | 0.5 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) |
| 0.2 | 4.1 | GO:0046697 | decidualization(GO:0046697) |
| 0.2 | 1.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.2 | 5.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 0.5 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.2 | 0.9 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.2 | 4.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.2 | 0.9 | GO:1903438 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.2 | 1.8 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.2 | 1.1 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 2.0 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.2 | 0.9 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.6 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 1.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.1 | GO:0009313 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.8 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.1 | 3.6 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 1.5 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 6.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 1.5 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 2.4 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.1 | 0.5 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.1 | 1.0 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.1 | 1.3 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.7 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.1 | 2.5 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 3.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.2 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.1 | 1.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.3 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.1 | 1.5 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 4.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 4.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.2 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 0.1 | 1.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.9 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 3.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 15.2 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 6.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.7 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 0.8 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 1.0 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.0 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.5 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 9.1 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 2.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 2.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 2.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.0 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 1.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 2.1 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.3 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 1.9 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.1 | 2.7 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.1 | 0.4 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.2 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 2.3 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 1.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.3 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.9 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 0.4 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.5 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.5 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.9 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.1 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 1.4 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.1 | 0.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.7 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 8.2 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 1.2 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.1 | 0.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.0 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 0.4 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 1.8 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 1.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.3 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.1 | 0.4 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 2.7 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 1.9 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 2.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 3.1 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.9 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.2 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 1.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.0 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 1.2 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) |
| 0.0 | 2.8 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.6 | GO:0046512 | sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.5 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 1.1 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 1.4 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.3 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 1.0 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 1.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 3.0 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 2.1 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.3 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.2 | GO:1904348 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) negative regulation of gastro-intestinal system smooth muscle contraction(GO:1904305) negative regulation of small intestine smooth muscle contraction(GO:1904348) |
| 0.0 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.5 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.7 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 1.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 3.7 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.6 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.5 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 1.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.0 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.6 | GO:0009583 | detection of light stimulus(GO:0009583) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 21.4 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 2.0 | 10.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.8 | 14.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.1 | 4.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 1.0 | 15.6 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.7 | 2.9 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.7 | 2.0 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.6 | 2.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.6 | 1.7 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.5 | 2.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 1.2 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.4 | 2.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 6.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 1.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.3 | 1.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 2.7 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 4.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 6.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.8 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.2 | 10.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 1.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 19.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 2.7 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 4.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 15.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 5.2 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 4.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 3.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 4.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 1.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 1.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 0.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.7 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.9 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.8 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 2.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 9.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 3.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 8.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 3.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 13.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 10.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.8 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.9 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.8 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.3 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.8 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 1.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 2.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.9 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 4.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 4.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 3.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 5.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.4 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 5.7 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.3 | 41.3 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 8.5 | 25.6 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 8.1 | 24.2 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 5.3 | 21.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 3.3 | 10.0 | GO:0046911 | metal chelating activity(GO:0046911) |
| 2.5 | 7.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 2.4 | 14.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 2.2 | 11.2 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 2.1 | 8.4 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 2.1 | 6.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.9 | 5.8 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 1.5 | 6.1 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 1.2 | 4.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 1.1 | 17.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.0 | 4.2 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 1.0 | 4.1 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.9 | 3.6 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.9 | 3.6 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.9 | 4.5 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.9 | 3.5 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.9 | 12.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.8 | 2.5 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.8 | 3.9 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.8 | 2.3 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.8 | 2.3 | GO:0004794 | L-serine ammonia-lyase activity(GO:0003941) L-threonine ammonia-lyase activity(GO:0004794) |
| 0.7 | 2.9 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.7 | 7.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.7 | 2.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.7 | 6.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.7 | 5.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.6 | 15.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.5 | 1.5 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.5 | 1.5 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.5 | 14.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 2.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.5 | 1.9 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.4 | 13.0 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.4 | 1.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.4 | 11.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.4 | 1.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 2.1 | GO:0030977 | taurine binding(GO:0030977) |
| 0.4 | 16.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 3.0 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.4 | 2.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.4 | 1.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.3 | 26.1 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.3 | 1.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 5.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 0.9 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.3 | 4.6 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.3 | 3.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.3 | 2.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 1.1 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.2 | 1.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.2 | 3.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 0.7 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.2 | 2.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 2.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 1.1 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 1.3 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 1.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 29.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 0.6 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.2 | 2.0 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.2 | 2.0 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 1.9 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 2.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 1.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 4.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 2.7 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.2 | 6.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 1.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 0.7 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 1.6 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 1.4 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 1.7 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 3.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 1.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 4.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 4.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 3.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 6.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 1.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 20.2 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 4.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.5 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 2.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 2.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 1.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.0 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.3 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.4 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.7 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.9 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 1.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.7 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 4.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 1.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 4.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.5 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.0 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 3.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.7 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 1.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 1.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 1.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 3.7 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 3.6 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 1.1 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.9 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 1.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 1.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 2.3 | GO:0005549 | odorant binding(GO:0005549) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 15.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 17.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 7.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 5.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 9.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 0.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 5.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.0 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 2.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 3.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 33.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.7 | 21.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.7 | 9.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.6 | 7.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.6 | 7.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.6 | 6.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.4 | 12.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.4 | 7.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 6.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 6.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 4.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 1.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.3 | 9.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 24.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 9.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 19.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 2.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 4.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 2.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.9 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 4.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 5.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 4.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 3.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 1.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 2.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.2 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |