Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
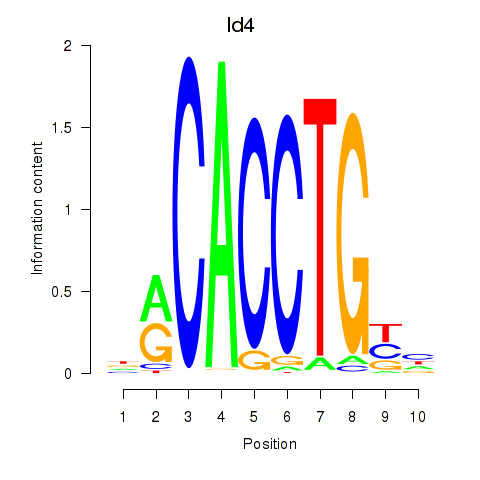
Results for Id4
Z-value: 2.74
Transcription factors associated with Id4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Id4
|
ENSMUSG00000021379.3 | inhibitor of DNA binding 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Id4 | mm39_v1_chr13_+_48414582_48414704 | -0.10 | 5.6e-01 | Click! |
Activity profile of Id4 motif
Sorted Z-values of Id4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_30623592 | 35.67 |
ENSMUST00000217812.2
ENSMUST00000074671.9 |
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr16_-_17906886 | 22.59 |
ENSMUST00000132241.2
ENSMUST00000139861.2 ENSMUST00000003620.13 |
Prodh
|
proline dehydrogenase |
| chr15_-_98575332 | 20.62 |
ENSMUST00000120997.2
ENSMUST00000109149.9 ENSMUST00000003451.11 |
Rnd1
|
Rho family GTPase 1 |
| chr4_+_133280680 | 20.28 |
ENSMUST00000042706.3
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr15_+_76582372 | 18.83 |
ENSMUST00000229140.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr10_+_87695352 | 18.07 |
ENSMUST00000121952.8
ENSMUST00000126490.8 |
Igf1
|
insulin-like growth factor 1 |
| chr11_-_5900019 | 15.49 |
ENSMUST00000102920.4
|
Gck
|
glucokinase |
| chr10_-_128796834 | 15.31 |
ENSMUST00000026398.5
|
Mettl7b
|
methyltransferase like 7B |
| chr7_+_43856724 | 15.07 |
ENSMUST00000077354.5
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chr11_+_72326391 | 14.80 |
ENSMUST00000100903.3
|
Ggt6
|
gamma-glutamyltransferase 6 |
| chr15_+_76579885 | 14.78 |
ENSMUST00000231028.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr7_+_140343652 | 14.67 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr15_+_76579960 | 13.99 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr2_+_153334710 | 13.44 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr14_-_47426863 | 13.18 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr7_+_127400016 | 13.12 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr7_-_30672747 | 12.77 |
ENSMUST00000205961.2
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr2_+_102489558 | 12.74 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr14_+_55813074 | 12.34 |
ENSMUST00000022826.7
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr7_-_30672824 | 12.29 |
ENSMUST00000147431.2
ENSMUST00000098553.11 ENSMUST00000108116.10 |
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr9_-_108444561 | 12.14 |
ENSMUST00000074208.6
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr7_-_30672889 | 12.04 |
ENSMUST00000001279.15
|
Lsr
|
lipolysis stimulated lipoprotein receptor |
| chr18_+_20691095 | 11.88 |
ENSMUST00000059787.15
ENSMUST00000120102.8 |
Dsg2
|
desmoglein 2 |
| chr3_-_67422821 | 11.87 |
ENSMUST00000054825.5
|
Rarres1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr1_-_91340884 | 11.81 |
ENSMUST00000086851.2
|
Hes6
|
hairy and enhancer of split 6 |
| chr10_+_87695886 | 11.80 |
ENSMUST00000062862.13
|
Igf1
|
insulin-like growth factor 1 |
| chr9_+_108174052 | 11.68 |
ENSMUST00000035230.7
|
Amt
|
aminomethyltransferase |
| chr10_+_87697155 | 11.65 |
ENSMUST00000122100.3
|
Igf1
|
insulin-like growth factor 1 |
| chr10_+_127637015 | 11.64 |
ENSMUST00000071646.2
|
Rdh16
|
retinol dehydrogenase 16 |
| chr2_+_31360219 | 11.51 |
ENSMUST00000102840.5
|
Ass1
|
argininosuccinate synthetase 1 |
| chr8_+_96078886 | 11.25 |
ENSMUST00000034243.7
|
Mmp15
|
matrix metallopeptidase 15 |
| chrX_+_100427331 | 10.59 |
ENSMUST00000119190.2
|
Gjb1
|
gap junction protein, beta 1 |
| chr14_-_31362909 | 10.33 |
ENSMUST00000022437.16
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr2_+_102488985 | 10.30 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr18_+_20691278 | 9.92 |
ENSMUST00000121837.2
|
Dsg2
|
desmoglein 2 |
| chr14_-_31362835 | 9.79 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr17_-_45997132 | 9.75 |
ENSMUST00000113523.9
|
Tmem63b
|
transmembrane protein 63b |
| chr10_+_127702326 | 9.62 |
ENSMUST00000092058.4
|
Rdh16f2
|
RDH16 family member 2 |
| chr19_-_40175709 | 9.57 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr9_+_77824646 | 9.40 |
ENSMUST00000034904.14
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr19_-_46661321 | 9.12 |
ENSMUST00000026012.8
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr14_-_57342150 | 8.91 |
ENSMUST00000055698.8
|
Gjb2
|
gap junction protein, beta 2 |
| chr8_-_41586713 | 8.70 |
ENSMUST00000155055.2
ENSMUST00000059115.13 ENSMUST00000145860.2 |
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr19_-_46661501 | 8.69 |
ENSMUST00000236174.2
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr2_-_103315483 | 8.60 |
ENSMUST00000028610.10
|
Cat
|
catalase |
| chr17_+_24971952 | 8.25 |
ENSMUST00000044922.8
ENSMUST00000234202.2 |
Hs3st6
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 6 |
| chr2_-_103315418 | 8.11 |
ENSMUST00000111168.4
|
Cat
|
catalase |
| chr18_+_84869456 | 8.11 |
ENSMUST00000160180.9
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr5_+_135015046 | 8.11 |
ENSMUST00000094245.4
|
Cldn3
|
claudin 3 |
| chr9_-_108443916 | 8.04 |
ENSMUST00000194381.2
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr2_+_119181703 | 7.89 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr2_-_91025208 | 7.81 |
ENSMUST00000111355.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr18_-_38345010 | 7.78 |
ENSMUST00000159405.3
ENSMUST00000160721.8 |
Pcdh1
|
protocadherin 1 |
| chr11_+_69983531 | 7.66 |
ENSMUST00000124721.2
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr11_-_120551494 | 7.62 |
ENSMUST00000106178.9
|
Notum
|
notum palmitoleoyl-protein carboxylesterase |
| chr14_+_21102642 | 7.59 |
ENSMUST00000045376.11
|
Adk
|
adenosine kinase |
| chr11_-_113600346 | 7.55 |
ENSMUST00000173655.8
ENSMUST00000100248.6 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr5_+_21391282 | 7.49 |
ENSMUST00000036031.13
ENSMUST00000198937.2 |
Gsap
|
gamma-secretase activating protein |
| chr2_+_71811526 | 7.49 |
ENSMUST00000090826.12
ENSMUST00000102698.10 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr11_+_102652228 | 7.44 |
ENSMUST00000103081.10
ENSMUST00000068150.7 |
Adam11
|
a disintegrin and metallopeptidase domain 11 |
| chr8_+_120121612 | 7.41 |
ENSMUST00000098367.5
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr2_-_121637505 | 7.40 |
ENSMUST00000138157.8
|
Frmd5
|
FERM domain containing 5 |
| chr16_-_45830575 | 7.38 |
ENSMUST00000130481.2
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr17_-_45997046 | 7.33 |
ENSMUST00000143907.3
ENSMUST00000127065.8 |
Tmem63b
|
transmembrane protein 63b |
| chr2_-_91025380 | 7.32 |
ENSMUST00000111356.8
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr7_-_28981335 | 7.23 |
ENSMUST00000108236.5
ENSMUST00000098604.12 |
Spint2
|
serine protease inhibitor, Kunitz type 2 |
| chr11_+_117700479 | 7.21 |
ENSMUST00000026649.14
ENSMUST00000177131.8 ENSMUST00000120928.2 ENSMUST00000175737.2 ENSMUST00000132298.2 |
Syngr2
Gm20708
|
synaptogyrin 2 predicted gene 20708 |
| chr15_+_99615396 | 7.20 |
ENSMUST00000023760.13
ENSMUST00000162194.2 |
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 (soluble) |
| chr14_+_21102662 | 7.20 |
ENSMUST00000223915.2
|
Adk
|
adenosine kinase |
| chr17_-_45996899 | 7.14 |
ENSMUST00000145873.8
|
Tmem63b
|
transmembrane protein 63b |
| chrX_+_10118544 | 7.13 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chr3_+_94280101 | 7.11 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr14_+_40827317 | 7.08 |
ENSMUST00000047286.7
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr2_-_91025441 | 7.06 |
ENSMUST00000002177.9
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr6_-_125357756 | 7.02 |
ENSMUST00000042647.7
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr17_-_35077089 | 6.94 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr9_-_21900724 | 6.87 |
ENSMUST00000045726.8
|
Rgl3
|
ral guanine nucleotide dissociation stimulator-like 3 |
| chrX_+_10118600 | 6.81 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr11_-_35871300 | 6.78 |
ENSMUST00000018993.7
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr4_-_137137088 | 6.69 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr2_-_37593287 | 6.68 |
ENSMUST00000072186.12
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr11_+_120421496 | 6.68 |
ENSMUST00000026119.8
|
Gcgr
|
glucagon receptor |
| chr15_-_100576715 | 6.68 |
ENSMUST00000229869.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr2_-_121637469 | 6.66 |
ENSMUST00000110592.2
|
Frmd5
|
FERM domain containing 5 |
| chr19_-_58443593 | 6.66 |
ENSMUST00000135730.2
ENSMUST00000152507.8 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr19_-_58443830 | 6.56 |
ENSMUST00000026076.14
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr10_-_24803336 | 6.53 |
ENSMUST00000020161.10
|
Arg1
|
arginase, liver |
| chr11_+_69983459 | 6.49 |
ENSMUST00000102572.8
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr18_+_24786748 | 6.41 |
ENSMUST00000068006.9
|
Mocos
|
molybdenum cofactor sulfurase |
| chr11_+_69983479 | 6.39 |
ENSMUST00000143772.8
|
Asgr2
|
asialoglycoprotein receptor 2 |
| chr7_-_48493388 | 6.37 |
ENSMUST00000167786.4
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr4_-_155445818 | 6.35 |
ENSMUST00000030922.15
|
Prkcz
|
protein kinase C, zeta |
| chr10_+_127595639 | 6.22 |
ENSMUST00000128247.2
|
Rdh16f1
|
RDH16 family member 1 |
| chr9_-_108141105 | 6.20 |
ENSMUST00000166905.8
ENSMUST00000191899.6 |
Dag1
|
dystroglycan 1 |
| chr8_+_36924702 | 6.20 |
ENSMUST00000135373.8
ENSMUST00000147525.9 |
Trmt9b
|
tRNA methyltransferase 9B |
| chr2_-_91025492 | 6.17 |
ENSMUST00000111354.2
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr5_+_35915217 | 6.16 |
ENSMUST00000101280.10
ENSMUST00000054598.12 ENSMUST00000114205.8 ENSMUST00000114206.9 |
Ablim2
|
actin-binding LIM protein 2 |
| chr9_+_37524966 | 6.15 |
ENSMUST00000215474.2
|
Siae
|
sialic acid acetylesterase |
| chr14_+_40827108 | 6.15 |
ENSMUST00000224514.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr15_+_88703786 | 6.11 |
ENSMUST00000024042.5
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr1_+_16175998 | 6.11 |
ENSMUST00000027053.8
|
Rdh10
|
retinol dehydrogenase 10 (all-trans) |
| chr11_-_67812960 | 6.09 |
ENSMUST00000021288.10
ENSMUST00000108677.2 |
Usp43
|
ubiquitin specific peptidase 43 |
| chr13_+_91889626 | 6.00 |
ENSMUST00000022120.5
|
Acot12
|
acyl-CoA thioesterase 12 |
| chrX_-_161426542 | 5.99 |
ENSMUST00000101102.2
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr8_+_77628916 | 5.94 |
ENSMUST00000109912.8
ENSMUST00000128862.2 ENSMUST00000109911.8 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr15_-_89263128 | 5.94 |
ENSMUST00000227834.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr2_-_65068960 | 5.93 |
ENSMUST00000112429.9
ENSMUST00000102726.8 ENSMUST00000112430.8 |
Cobll1
|
Cobl-like 1 |
| chr18_-_35760260 | 5.91 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr1_+_171246593 | 5.90 |
ENSMUST00000171362.2
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr9_-_43151179 | 5.89 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr10_+_75729237 | 5.89 |
ENSMUST00000009236.6
ENSMUST00000217811.2 |
Derl3
|
Der1-like domain family, member 3 |
| chr9_+_53212871 | 5.84 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr6_-_127086480 | 5.80 |
ENSMUST00000039913.9
|
Tigar
|
Trp53 induced glycolysis regulatory phosphatase |
| chr11_-_120675009 | 5.77 |
ENSMUST00000026156.8
|
Rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr9_+_102885156 | 5.77 |
ENSMUST00000035148.13
|
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr6_-_119521243 | 5.77 |
ENSMUST00000119369.2
ENSMUST00000178696.8 |
Wnt5b
|
wingless-type MMTV integration site family, member 5B |
| chr11_-_50216426 | 5.75 |
ENSMUST00000179865.8
ENSMUST00000020637.9 |
Canx
|
calnexin |
| chr2_+_26969384 | 5.70 |
ENSMUST00000091233.7
|
Adamtsl2
|
ADAMTS-like 2 |
| chr15_+_54434576 | 5.66 |
ENSMUST00000025356.4
|
Mal2
|
mal, T cell differentiation protein 2 |
| chr12_+_113106407 | 5.61 |
ENSMUST00000196015.5
|
Crip2
|
cysteine rich protein 2 |
| chrX_+_35375751 | 5.56 |
ENSMUST00000033418.8
|
Il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr2_-_65068917 | 5.55 |
ENSMUST00000090896.10
ENSMUST00000155082.2 |
Cobll1
|
Cobl-like 1 |
| chr17_+_37253802 | 5.54 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr7_+_143027473 | 5.54 |
ENSMUST00000052348.12
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr7_+_127399776 | 5.52 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr14_+_40826970 | 5.51 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr4_+_139350152 | 5.51 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr16_+_31482658 | 5.47 |
ENSMUST00000115201.8
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr8_-_106660470 | 5.47 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr15_-_83054369 | 5.43 |
ENSMUST00000162834.3
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr7_+_127399789 | 5.43 |
ENSMUST00000125188.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr1_+_182591425 | 5.42 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr6_+_125298372 | 5.39 |
ENSMUST00000176442.8
ENSMUST00000177329.2 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr11_-_113600838 | 5.38 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr17_+_29077385 | 5.37 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr9_-_108140925 | 5.37 |
ENSMUST00000171412.7
ENSMUST00000195429.6 ENSMUST00000080435.9 |
Dag1
|
dystroglycan 1 |
| chr10_-_126956991 | 5.33 |
ENSMUST00000080975.6
ENSMUST00000164259.9 |
Os9
|
amplified in osteosarcoma |
| chr16_-_4950285 | 5.31 |
ENSMUST00000035672.5
|
Ppl
|
periplakin |
| chr7_-_27037096 | 5.29 |
ENSMUST00000038618.13
ENSMUST00000108369.9 |
Ltbp4
|
latent transforming growth factor beta binding protein 4 |
| chr18_+_32064358 | 5.29 |
ENSMUST00000025254.9
|
Lims2
|
LIM and senescent cell antigen like domains 2 |
| chr4_-_104967032 | 5.29 |
ENSMUST00000030243.8
|
Prkaa2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit |
| chr15_+_25843225 | 5.22 |
ENSMUST00000022881.15
|
Retreg1
|
reticulophagy regulator 1 |
| chr17_-_32639936 | 5.21 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr18_-_62044871 | 5.20 |
ENSMUST00000166783.3
ENSMUST00000049378.15 |
Ablim3
|
actin binding LIM protein family, member 3 |
| chr17_+_26036893 | 5.19 |
ENSMUST00000235694.2
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr7_+_96981517 | 5.19 |
ENSMUST00000054107.6
|
Kctd21
|
potassium channel tetramerisation domain containing 21 |
| chr7_+_65511482 | 5.15 |
ENSMUST00000176199.8
|
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr8_+_56747613 | 5.15 |
ENSMUST00000034026.10
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD) |
| chr17_-_57023788 | 5.11 |
ENSMUST00000067931.7
|
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr15_-_89263790 | 5.11 |
ENSMUST00000238996.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr2_+_25318642 | 5.09 |
ENSMUST00000102919.4
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr18_-_16942289 | 5.03 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2 |
| chr11_-_20781009 | 5.02 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr11_-_12414850 | 4.98 |
ENSMUST00000109650.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr15_-_89263448 | 4.98 |
ENSMUST00000049968.9
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr4_-_96441854 | 4.94 |
ENSMUST00000030303.6
|
Cyp2j6
|
cytochrome P450, family 2, subfamily j, polypeptide 6 |
| chr7_+_127399848 | 4.93 |
ENSMUST00000139068.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr10_-_81243475 | 4.92 |
ENSMUST00000140916.8
|
Nfic
|
nuclear factor I/C |
| chr16_+_31482745 | 4.87 |
ENSMUST00000100001.10
ENSMUST00000064477.14 |
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr5_-_25305621 | 4.84 |
ENSMUST00000030784.14
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr14_+_31363004 | 4.82 |
ENSMUST00000090147.7
|
Btd
|
biotinidase |
| chr9_-_43117052 | 4.81 |
ENSMUST00000176636.5
|
Pou2f3
|
POU domain, class 2, transcription factor 3 |
| chr2_+_126394327 | 4.81 |
ENSMUST00000061491.14
|
Slc27a2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr17_-_45906428 | 4.79 |
ENSMUST00000171081.8
ENSMUST00000172301.8 ENSMUST00000167332.8 ENSMUST00000170488.8 ENSMUST00000167195.8 ENSMUST00000064889.13 ENSMUST00000051574.13 ENSMUST00000164217.8 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr11_-_100661762 | 4.78 |
ENSMUST00000139341.2
ENSMUST00000017891.14 |
Ghdc
|
GH3 domain containing |
| chr1_-_184615415 | 4.77 |
ENSMUST00000048308.6
|
C130074G19Rik
|
RIKEN cDNA C130074G19 gene |
| chr1_+_135746330 | 4.77 |
ENSMUST00000038760.10
|
Lad1
|
ladinin |
| chr11_-_43727071 | 4.75 |
ENSMUST00000167574.2
|
Adra1b
|
adrenergic receptor, alpha 1b |
| chr7_-_80051455 | 4.75 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr6_+_134807097 | 4.66 |
ENSMUST00000046303.12
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr18_+_74575623 | 4.65 |
ENSMUST00000121875.8
|
Myo5b
|
myosin VB |
| chr7_-_99276310 | 4.65 |
ENSMUST00000178124.3
|
Tpbgl
|
trophoblast glycoprotein-like |
| chr19_-_58442866 | 4.61 |
ENSMUST00000169850.8
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr3_+_94269745 | 4.59 |
ENSMUST00000169433.3
|
C2cd4d
|
C2 calcium-dependent domain containing 4D |
| chr5_+_35915290 | 4.57 |
ENSMUST00000114204.8
ENSMUST00000129347.8 |
Ablim2
|
actin-binding LIM protein 2 |
| chr13_-_100152069 | 4.56 |
ENSMUST00000022148.7
|
Mccc2
|
methylcrotonoyl-Coenzyme A carboxylase 2 (beta) |
| chr18_+_74575567 | 4.55 |
ENSMUST00000074157.13
|
Myo5b
|
myosin VB |
| chr1_+_192835414 | 4.55 |
ENSMUST00000076521.7
|
Irf6
|
interferon regulatory factor 6 |
| chr3_-_89294430 | 4.54 |
ENSMUST00000107433.8
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr11_-_100418717 | 4.54 |
ENSMUST00000107389.8
ENSMUST00000007131.16 |
Acly
|
ATP citrate lyase |
| chr4_-_155445779 | 4.49 |
ENSMUST00000105624.2
|
Prkcz
|
protein kinase C, zeta |
| chr6_+_113460258 | 4.46 |
ENSMUST00000032422.6
|
Creld1
|
cysteine-rich with EGF-like domains 1 |
| chr2_-_64853083 | 4.46 |
ENSMUST00000028252.14
|
Grb14
|
growth factor receptor bound protein 14 |
| chr5_+_138170259 | 4.43 |
ENSMUST00000019662.11
ENSMUST00000151318.8 |
Ap4m1
|
adaptor-related protein complex AP-4, mu 1 |
| chr7_+_119499322 | 4.43 |
ENSMUST00000106516.2
|
Lyrm1
|
LYR motif containing 1 |
| chr7_+_44033520 | 4.41 |
ENSMUST00000118962.8
ENSMUST00000118831.8 |
Syt3
|
synaptotagmin III |
| chr19_-_30152814 | 4.41 |
ENSMUST00000025778.9
|
Gldc
|
glycine decarboxylase |
| chr4_-_135221810 | 4.38 |
ENSMUST00000105856.9
|
Nipal3
|
NIPA-like domain containing 3 |
| chr11_-_119438569 | 4.36 |
ENSMUST00000026670.5
|
Nptx1
|
neuronal pentraxin 1 |
| chr8_-_84738761 | 4.35 |
ENSMUST00000191523.2
ENSMUST00000190457.2 ENSMUST00000185457.2 |
Misp3
|
MISP family member 3 |
| chr6_-_47790272 | 4.35 |
ENSMUST00000077290.9
|
Pdia4
|
protein disulfide isomerase associated 4 |
| chr9_+_80072274 | 4.34 |
ENSMUST00000035889.15
ENSMUST00000113268.8 |
Myo6
|
myosin VI |
| chrX_+_21581135 | 4.31 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr9_-_108140904 | 4.29 |
ENSMUST00000194698.2
|
Dag1
|
dystroglycan 1 |
| chr11_-_51647204 | 4.29 |
ENSMUST00000109092.8
ENSMUST00000064297.5 |
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr7_-_127545896 | 4.28 |
ENSMUST00000118755.8
ENSMUST00000094026.10 |
Prss36
|
protease, serine 36 |
| chr13_-_90237713 | 4.27 |
ENSMUST00000022115.14
|
Xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr12_+_113103817 | 4.27 |
ENSMUST00000084882.9
|
Crip2
|
cysteine rich protein 2 |
| chr16_+_38722666 | 4.26 |
ENSMUST00000023478.8
|
Igsf11
|
immunoglobulin superfamily, member 11 |
| chr1_+_182591771 | 4.26 |
ENSMUST00000193660.6
|
Susd4
|
sushi domain containing 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Id4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.4 | 37.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 9.4 | 28.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 8.9 | 35.7 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 7.3 | 21.8 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 7.1 | 28.4 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 6.2 | 18.7 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 5.9 | 41.5 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 5.4 | 16.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 5.3 | 15.9 | GO:0021682 | nerve maturation(GO:0021682) |
| 4.9 | 14.8 | GO:0006175 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 4.6 | 13.9 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 3.8 | 18.9 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 3.6 | 24.9 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 3.4 | 13.5 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 3.3 | 13.2 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 3.3 | 23.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 3.2 | 3.2 | GO:1904173 | regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 3.0 | 8.9 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 2.9 | 11.5 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 2.6 | 29.0 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 2.5 | 7.6 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 2.5 | 7.4 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 2.4 | 9.7 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 2.2 | 6.7 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 2.1 | 6.4 | GO:1903920 | positive regulation of actin filament severing(GO:1903920) |
| 2.1 | 6.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 2.0 | 4.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 2.0 | 11.9 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 2.0 | 9.9 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 2.0 | 9.9 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.9 | 5.8 | GO:1902689 | negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 1.7 | 6.8 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 1.7 | 23.6 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 1.7 | 9.9 | GO:0032439 | endosome localization(GO:0032439) |
| 1.6 | 4.9 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 1.6 | 4.8 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 1.5 | 10.8 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 1.5 | 4.6 | GO:0034226 | nitric oxide production involved in inflammatory response(GO:0002537) lysine import(GO:0034226) L-lysine import(GO:0061461) L-ornithine transmembrane transport(GO:1903352) L-arginine transmembrane transport(GO:1903400) L-lysine transmembrane transport(GO:1903401) L-lysine import into cell(GO:1903410) arginine transmembrane transport(GO:1903826) |
| 1.5 | 6.0 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 1.5 | 4.5 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 1.5 | 5.9 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 1.5 | 11.7 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 1.4 | 4.3 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 1.4 | 5.6 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 1.4 | 4.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 1.3 | 11.6 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.3 | 9.0 | GO:0015862 | uridine transport(GO:0015862) |
| 1.3 | 10.1 | GO:0000050 | urea cycle(GO:0000050) |
| 1.3 | 3.8 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 1.2 | 3.7 | GO:0019085 | early viral transcription(GO:0019085) |
| 1.2 | 8.7 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 1.2 | 8.6 | GO:0001757 | somite specification(GO:0001757) |
| 1.2 | 6.0 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 1.2 | 3.6 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 1.2 | 4.8 | GO:0007113 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.2 | 6.0 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.2 | 8.3 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 1.2 | 10.7 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 1.2 | 12.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 1.2 | 9.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.1 | 4.5 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 1.1 | 12.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 1.1 | 3.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 1.1 | 7.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.1 | 2.2 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 1.1 | 8.6 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 1.0 | 19.3 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 1.0 | 11.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 1.0 | 3.0 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.0 | 4.0 | GO:0015755 | fructose transport(GO:0015755) |
| 1.0 | 3.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 1.0 | 14.8 | GO:0009650 | UV protection(GO:0009650) |
| 1.0 | 9.8 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 1.0 | 9.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.0 | 10.6 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.9 | 2.8 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.9 | 20.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.9 | 10.5 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.9 | 3.5 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.9 | 2.6 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.9 | 6.0 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.8 | 6.7 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.8 | 4.9 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.8 | 1.6 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.8 | 2.3 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.7 | 1.5 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.7 | 3.7 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.7 | 2.2 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) oxalic acid secretion(GO:0046724) |
| 0.7 | 10.9 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.7 | 7.9 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.7 | 2.8 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.7 | 7.1 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.7 | 8.3 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.7 | 22.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.7 | 2.1 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.7 | 6.2 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.7 | 2.0 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.7 | 2.0 | GO:0071603 | trabecular meshwork development(GO:0002930) dibenzo-p-dioxin metabolic process(GO:0018894) endothelial cell-cell adhesion(GO:0071603) |
| 0.7 | 4.6 | GO:0035865 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.6 | 3.9 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.6 | 4.5 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.6 | 1.9 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.6 | 3.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.6 | 3.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) negative regulation of serotonin secretion(GO:0014063) |
| 0.6 | 3.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.6 | 4.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.6 | 6.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.6 | 1.2 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.6 | 3.0 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.6 | 3.5 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.6 | 4.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.6 | 2.9 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.6 | 5.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.6 | 1.7 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.6 | 8.6 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.6 | 3.4 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.6 | 2.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.6 | 1.7 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.6 | 1.7 | GO:0019230 | proprioception(GO:0019230) |
| 0.6 | 6.7 | GO:0007567 | parturition(GO:0007567) |
| 0.6 | 3.3 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.5 | 2.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.5 | 3.8 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.5 | 5.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.5 | 4.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.5 | 3.7 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.5 | 25.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.5 | 5.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.5 | 2.6 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.5 | 2.1 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.5 | 2.6 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.5 | 8.2 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.5 | 7.6 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.5 | 1.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.5 | 2.0 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.5 | 2.0 | GO:0019323 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) pentose catabolic process(GO:0019323) |
| 0.5 | 4.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.5 | 1.5 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.5 | 2.4 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.5 | 6.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.5 | 1.8 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.5 | 3.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.5 | 1.4 | GO:0060061 | Spemann organizer formation(GO:0060061) syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.4 | 12.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.4 | 3.6 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.4 | 1.8 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.4 | 7.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.4 | 2.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.4 | 7.4 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.4 | 4.8 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.4 | 3.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.4 | 1.7 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.4 | 3.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.4 | 6.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.4 | 2.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.4 | 2.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.4 | 2.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.4 | 10.0 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.4 | 1.2 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.4 | 2.9 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.4 | 1.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.4 | 2.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.4 | 0.8 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.4 | 4.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.4 | 2.0 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.4 | 5.2 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.4 | 2.8 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.4 | 4.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.4 | 4.3 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.4 | 1.6 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.4 | 1.2 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.4 | 6.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.4 | 6.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.4 | 3.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.4 | 0.8 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.4 | 7.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.4 | 4.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 | 3.0 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.4 | 1.5 | GO:0070268 | cornification(GO:0070268) |
| 0.4 | 2.3 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.4 | 1.5 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.4 | 0.7 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.4 | 4.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.4 | 1.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.4 | 2.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.4 | 1.8 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.4 | 3.3 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.4 | 0.7 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.4 | 3.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.4 | 1.4 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.4 | 0.4 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.4 | 8.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 1.1 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.4 | 6.0 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.4 | 2.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.3 | 5.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.3 | 1.7 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.3 | 1.4 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.3 | 1.0 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.3 | 3.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.3 | 3.4 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.3 | 1.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.3 | 1.0 | GO:2000832 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) negative regulation of steroid hormone secretion(GO:2000832) |
| 0.3 | 3.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.3 | 5.0 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.3 | 5.0 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.3 | 5.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 1.3 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.3 | 2.9 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.3 | 2.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.3 | 1.0 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.3 | 4.2 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.3 | 1.6 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.3 | 4.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.3 | 4.4 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 2.5 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.3 | 0.9 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.3 | 8.1 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.3 | 11.1 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.3 | 1.5 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.3 | 6.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.3 | 1.8 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.3 | 5.5 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.3 | 7.3 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.3 | 2.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.3 | 2.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.3 | 7.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.3 | 4.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 2.0 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.3 | 0.6 | GO:2000849 | glucocorticoid secretion(GO:0035933) cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) positive regulation of cortisol secretion(GO:0051464) regulation of glucocorticoid secretion(GO:2000849) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.3 | 0.8 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.3 | 0.6 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.3 | 1.7 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.3 | 1.7 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.3 | 0.5 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.3 | 9.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.3 | 1.1 | GO:0048104 | embryonic nail plate morphogenesis(GO:0035880) establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.3 | 0.8 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.3 | 3.5 | GO:2000191 | regulation of fatty acid transport(GO:2000191) |
| 0.3 | 3.0 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.3 | 3.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.3 | 5.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.3 | 1.6 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.3 | 0.8 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.3 | 1.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.2 | 0.7 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 | 0.7 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 1.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 2.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 3.7 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.2 | 0.5 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 | 0.7 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.2 | 1.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 5.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 0.7 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 2.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 8.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 2.9 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.2 | 2.6 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.2 | 3.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 1.4 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.2 | 10.0 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.2 | 3.5 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.2 | 0.7 | GO:0043366 | beta selection(GO:0043366) |
| 0.2 | 20.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.2 | 22.9 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.2 | 3.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 0.9 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.2 | 0.9 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 0.5 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.2 | 4.9 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.2 | 4.4 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.2 | 2.0 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.2 | 1.5 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 7.6 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.2 | 0.4 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.2 | 0.6 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.2 | 0.4 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.2 | 2.4 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.2 | 1.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 0.8 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.2 | 0.6 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.2 | 6.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.2 | 0.6 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.2 | 2.0 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 0.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 0.2 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.2 | 0.6 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.2 | 3.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 1.6 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 2.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.2 | 0.8 | GO:0090282 | trophectodermal cellular morphogenesis(GO:0001831) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.2 | 1.9 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.2 | 2.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 0.8 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 2.4 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 1.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.2 | 2.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 0.9 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 | 0.7 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.2 | 6.5 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.2 | 1.5 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.2 | 2.4 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.2 | 1.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 6.9 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.2 | 1.6 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.2 | 0.9 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 3.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 0.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 3.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 1.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.2 | 0.5 | GO:0048627 | myoblast development(GO:0048627) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 | 0.2 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.2 | 5.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 0.7 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 1.5 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.2 | 2.0 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.2 | 1.5 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.2 | 1.5 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.2 | 7.7 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 0.8 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.2 | 1.3 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.2 | 1.8 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.2 | 1.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.2 | 3.1 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.2 | 0.5 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 0.8 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.2 | 4.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.2 | 1.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 6.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.2 | 0.6 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 3.6 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 0.5 | GO:0021679 | cerebellar molecular layer development(GO:0021679) vestibular nucleus development(GO:0021750) |
| 0.2 | 4.8 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.2 | 1.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 1.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.2 | 2.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 0.6 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.1 | 4.6 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 | 0.9 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 7.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 1.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.4 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 2.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 1.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 1.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.6 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 1.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.6 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 5.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.8 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 1.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 4.7 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 6.0 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 0.6 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 1.8 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 2.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 1.4 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.1 | 4.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 2.5 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 1.7 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 3.4 | GO:0060749 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.1 | 0.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 5.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.8 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 9.9 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 1.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 5.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.1 | 0.2 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.1 | 5.8 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 0.7 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 1.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 2.3 | GO:0010863 | activation of phospholipase C activity(GO:0007202) positive regulation of phospholipase C activity(GO:0010863) |
| 0.1 | 0.7 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 2.8 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.5 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 1.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.3 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.1 | 3.0 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.1 | 0.2 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 0.1 | 0.7 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.0 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.1 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.2 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.1 | 1.0 | GO:0045617 | negative regulation of epidermal cell differentiation(GO:0045605) negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 4.1 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 1.6 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 2.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 2.0 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.4 | GO:1902477 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 2.2 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 1.5 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.5 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 | 2.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 4.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 2.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.2 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 4.7 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.6 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.1 | 0.2 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.1 | 0.2 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.3 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 2.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.0 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.7 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.1 | 0.5 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 3.4 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.1 | 0.5 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.1 | 1.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 2.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.7 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.2 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 4.2 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 0.8 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.1 | 1.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.5 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 3.9 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.1 | 1.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.3 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.6 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 0.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 1.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.4 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.1 | 0.6 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.4 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 | 2.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.4 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 0.4 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.0 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.4 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 2.3 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 1.2 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.1 | 0.6 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0070346 | white fat cell proliferation(GO:0070343) positive regulation of fat cell proliferation(GO:0070346) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 1.1 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.8 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 1.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.6 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 2.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 8.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.5 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 2.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 2.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 2.5 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.3 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.6 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.7 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.3 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 3.2 | GO:0048736 | appendage development(GO:0048736) limb development(GO:0060173) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 1.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 1.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 1.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 1.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.8 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.8 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 3.2 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.8 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.2 | GO:0045141 | telomere localization(GO:0034397) meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 | 0.2 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.3 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 2.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.7 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 5.0 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 0.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 1.3 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.4 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 2.7 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.3 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.5 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.0 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 1.2 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.2 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 42.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 3.6 | 32.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 2.4 | 9.4 | GO:0097447 | dendritic tree(GO:0097447) |
| 1.8 | 20.1 | GO:0045179 | apical cortex(GO:0045179) |
| 1.8 | 10.9 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 1.8 | 8.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.7 | 13.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.6 | 4.9 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 1.5 | 7.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.3 | 16.2 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 1.2 | 3.6 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 1.2 | 8.1 | GO:0070695 | FHF complex(GO:0070695) |
| 1.2 | 1.2 | GO:0019866 | organelle inner membrane(GO:0019866) |
| 1.1 | 8.5 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 1.0 | 18.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.0 | 29.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.9 | 6.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.9 | 22.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.8 | 3.4 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.8 | 15.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.8 | 4.0 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.7 | 3.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.7 | 2.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.6 | 10.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.6 | 1.9 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.6 | 12.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.6 | 1.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.6 | 9.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.6 | 29.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.6 | 7.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.6 | 8.9 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.5 | 4.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.5 | 1.5 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.5 | 8.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.5 | 4.0 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.5 | 5.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.5 | 13.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.5 | 2.8 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.4 | 5.8 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.4 | 9.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.4 | 1.7 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.4 | 7.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.4 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 3.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.4 | 5.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 4.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.4 | 1.8 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.4 | 3.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.3 | 10.8 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.3 | 10.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 1.0 | GO:0060473 | cortical granule(GO:0060473) |
| 0.3 | 1.6 | GO:0043293 | apoptosome(GO:0043293) |
| 0.3 | 1.5 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 6.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 17.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.3 | 0.8 | GO:0036501 | UFD1-NPL4 complex(GO:0036501) |
| 0.3 | 4.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 2.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 2.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 7.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.3 | 22.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 2.6 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.2 | 2.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 1.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 0.9 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 1.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 1.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 3.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 2.6 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 6.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 6.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 2.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 4.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 1.0 | GO:0043219 | lateral loop(GO:0043219) |
| 0.2 | 1.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 1.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 36.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.2 | 5.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 2.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 8.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 2.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 0.6 | GO:1904602 | serotonin-activated cation-selective channel complex(GO:1904602) |
| 0.2 | 6.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 12.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 3.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.2 | 1.7 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 5.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 2.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 5.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 2.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 1.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 1.7 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.2 | 2.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 11.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 64.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 1.5 | GO:0070578 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.1 | 5.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 6.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 18.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 2.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 5.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 1.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 7.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 28.6 | GO:0005740 | mitochondrial envelope(GO:0005740) |
| 0.1 | 18.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 2.9 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 5.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.6 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 4.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 4.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 3.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 2.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.8 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.5 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 3.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 21.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.7 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.3 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 4.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.8 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 0.8 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 2.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 2.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 20.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 7.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 0.3 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.3 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 3.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.6 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.1 | 3.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 12.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 22.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 2.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 2.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 9.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 3.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 53.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 2.2 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 1.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 5.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.0 | 0.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 3.5 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.3 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.5 | 47.6 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 7.2 | 29.0 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 5.3 | 31.9 | GO:0032810 | sterol response element binding(GO:0032810) |
| 4.9 | 14.8 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 4.7 | 18.7 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 4.2 | 16.7 | GO:0004096 | catalase activity(GO:0004096) |
| 3.8 | 23.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 3.5 | 13.9 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 2.5 | 7.6 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 2.4 | 7.2 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 2.2 | 6.7 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 2.1 | 6.3 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 2.0 | 12.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 2.0 | 15.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 2.0 | 5.9 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 1.9 | 13.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.9 | 7.6 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.7 | 20.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 1.6 | 9.9 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.5 | 15.5 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 1.5 | 4.6 | GO:0005287 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 1.5 | 9.0 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 1.5 | 4.5 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 1.5 | 13.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 1.5 | 37.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 1.4 | 5.7 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.4 | 12.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 1.3 | 4.0 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) uniporter activity(GO:0015292) |
| 1.3 | 7.9 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 1.3 | 15.7 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 1.3 | 6.4 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 1.3 | 6.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.3 | 20.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 1.2 | 4.9 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 1.2 | 8.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.2 | 3.7 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 1.2 | 9.8 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 1.2 | 6.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.2 | 6.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.2 | 4.8 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 1.2 | 7.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.1 | 4.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 1.1 | 11.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 1.0 | 4.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 1.0 | 24.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 1.0 | 3.0 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 1.0 | 44.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 1.0 | 3.0 | GO:0004320 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) S-acetyltransferase activity(GO:0016418) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) phosphopantetheine binding(GO:0031177) |
| 1.0 | 4.8 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.9 | 6.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.9 | 6.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.9 | 10.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.9 | 10.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.9 | 3.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.9 | 3.4 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.8 | 17.8 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.8 | 1.7 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.8 | 3.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.8 | 4.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.8 | 4.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.8 | 2.4 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.8 | 3.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.8 | 4.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.8 | 2.3 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.7 | 14.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.7 | 28.1 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.7 | 13.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.7 | 24.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.7 | 4.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.7 | 2.0 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.7 | 2.0 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.7 | 10.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.6 | 9.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 3.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.6 | 1.8 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.6 | 2.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.6 | 5.0 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.5 | 2.2 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.5 | 1.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.5 | 4.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.5 | 2.2 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.5 | 2.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.5 | 1.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.5 | 2.0 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.5 | 3.6 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.5 | 3.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.5 | 2.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 3.0 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.5 | 9.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.5 | 2.4 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.5 | 1.9 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.5 | 1.9 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.5 | 1.4 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.5 | 2.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.5 | 1.4 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.5 | 1.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 11.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.5 | 4.6 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.5 | 5.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.5 | 2.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.4 | 3.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 3.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.4 | 1.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.4 | 0.9 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.4 | 1.3 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.4 | 2.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.4 | 10.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.4 | 4.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.4 | 1.6 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.4 | 10.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.4 | 1.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.4 | 3.5 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.4 | 4.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 2.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 2.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.4 | 2.6 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.4 | 5.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.4 | 2.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 1.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.4 | 1.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.4 | 1.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 5.3 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.3 | 1.4 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.3 | 2.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.3 | 1.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.3 | 2.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.3 | 9.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.3 | 1.0 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.3 | 6.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 3.9 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.3 | 3.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 15.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.3 | 6.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 0.9 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.3 | 6.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 5.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 7.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 5.5 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.3 | 15.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 1.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.3 | 3.9 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 3.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.3 | 4.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 1.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 0.8 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.3 | 1.4 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.3 | 1.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.3 | 8.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 11.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.3 | 1.1 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.3 | 31.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.3 | 1.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 0.8 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.3 | 1.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.3 | 1.3 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.2 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 5.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.7 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.2 | 5.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 4.6 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 0.7 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 3.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 8.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 6.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 1.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 1.2 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 6.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 2.7 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 11.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.2 | 1.8 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 2.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 1.7 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 2.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 2.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 0.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 4.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 0.6 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 0.4 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.2 | 0.6 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.2 | 3.6 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.2 | 1.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 1.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 5.5 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.2 | 0.6 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.2 | 2.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 1.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 0.7 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 13.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 3.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 3.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 3.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 2.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 3.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 4.4 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.2 | 1.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.2 | 1.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 1.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 1.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 3.0 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.2 | 0.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 1.0 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 8.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 6.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 1.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 2.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 2.6 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 30.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 1.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 6.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 2.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 7.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 16.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 0.9 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 3.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.4 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 5.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 3.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.8 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.1 | 4.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 4.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.0 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 4.9 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 7.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 34.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 2.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.4 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.1 | 1.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.6 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 1.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 1.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.6 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 6.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 1.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 4.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.3 | GO:1905030 | voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 4.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 3.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 0.6 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 3.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 7.5 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 1.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.8 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 2.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 5.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 4.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 5.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 3.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.3 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 0.2 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.2 | GO:1904315 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 9.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 2.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 7.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.1 | 0.6 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 3.3 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 1.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 3.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 4.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 2.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 3.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 3.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 5.3 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 2.0 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 12.1 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.1 | 2.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.8 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 1.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 6.0 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 2.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 3.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 4.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.6 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 21.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 12.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.5 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 1.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 3.0 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.0 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.5 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.0 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 1.0 | 20.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.8 | 5.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.7 | 49.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.6 | 34.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.5 | 14.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.5 | 41.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.4 | 11.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 4.9 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.3 | 4.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 4.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 8.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.3 | 4.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 10.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.3 | 12.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.2 | 10.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 7.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 5.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 5.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 5.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 12.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 11.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 7.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 11.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 2.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 12.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 5.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 3.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 7.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 8.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 2.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 5.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 6.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 3.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 1.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 3.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 3.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 0.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 15.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 1.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 6.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 14.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 13.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 7.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.6 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 2.3 | 33.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.8 | 38.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.6 | 47.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 1.6 | 20.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 1.1 | 22.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 1.0 | 20.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.9 | 16.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.7 | 10.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.7 | 11.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.7 | 12.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.7 | 7.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.7 | 9.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.7 | 70.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.6 | 19.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.6 | 6.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.6 | 20.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.6 | 20.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.5 | 3.1 | REACTOME SIGNALLING TO RAS | Genes involved in Signalling to RAS |
| 0.5 | 6.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.5 | 6.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.5 | 13.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.5 | 8.2 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.5 | 0.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.5 | 9.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.5 | 5.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 10.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 13.0 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.4 | 4.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.4 | 5.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 5.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 30.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.4 | 10.9 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.4 | 5.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 14.2 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.3 | 3.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 14.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 8.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 65.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.3 | 7.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 8.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 2.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 2.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 3.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 4.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 5.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 3.6 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.3 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 5.0 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.3 | 16.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 7.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 9.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 9.8 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.2 | 3.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 6.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 5.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 3.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 5.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 6.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 3.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 3.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 3.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 2.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 2.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 10.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 3.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 1.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 5.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 6.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 2.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 3.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 7.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 4.2 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 1.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 2.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 3.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 3.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 3.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 2.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 7.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.9 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 3.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 1.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 4.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.0 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR MUTANTS | Genes involved in Signaling by FGFR mutants |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 1.4 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.8 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.5 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 1.2 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME PKB MEDIATED EVENTS | Genes involved in PKB-mediated events |
| 0.0 | 1.0 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |