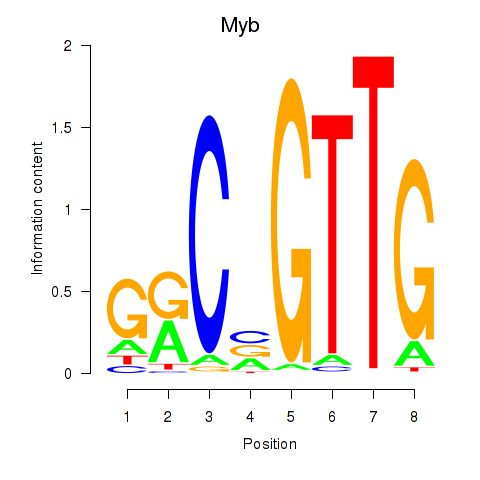
|
chr6_+_86055018
Show fit
|
29.06 |
ENSMUST00000205034.3
ENSMUST00000203724.3
|
Add2
|
adducin 2 (beta)
|
|
chr17_-_48758538
Show fit
|
28.81 |
ENSMUST00000024794.12
|
Tspo2
|
translocator protein 2
|
|
chr3_+_146110387
Show fit
|
27.88 |
ENSMUST00000106151.8
ENSMUST00000106153.9
ENSMUST00000039021.11
ENSMUST00000106149.8
ENSMUST00000149262.8
|
Ssx2ip
|
synovial sarcoma, X 2 interacting protein
|
|
chr10_+_79715448
Show fit
|
23.90 |
ENSMUST00000006679.15
|
Prtn3
|
proteinase 3
|
|
chr6_+_86055048
Show fit
|
23.69 |
ENSMUST00000032069.8
|
Add2
|
adducin 2 (beta)
|
|
chr4_-_118294521
Show fit
|
22.96 |
ENSMUST00000006565.13
|
Cdc20
|
cell division cycle 20
|
|
chr11_+_87684299
Show fit
|
21.19 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase
|
|
chr6_-_125168637
Show fit
|
20.45 |
ENSMUST00000043848.11
|
Ncapd2
|
non-SMC condensin I complex, subunit D2
|
|
chr6_+_124806506
Show fit
|
20.04 |
ENSMUST00000150120.8
|
Cdca3
|
cell division cycle associated 3
|
|
chr6_+_124806541
Show fit
|
19.52 |
ENSMUST00000024270.14
|
Cdca3
|
cell division cycle associated 3
|
|
chr9_+_72345801
Show fit
|
18.17 |
ENSMUST00000184604.8
ENSMUST00000034746.10
|
Mns1
|
meiosis-specific nuclear structural protein 1
|
|
chr17_-_34109513
Show fit
|
17.87 |
ENSMUST00000173386.2
ENSMUST00000114361.9
ENSMUST00000173492.9
|
Kifc1
|
kinesin family member C1
|
|
chr1_-_131066004
Show fit
|
17.48 |
ENSMUST00000016670.9
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3
|
|
chr6_-_131365380
Show fit
|
17.14 |
ENSMUST00000032309.13
ENSMUST00000087865.4
|
Ybx3
|
Y box protein 3
|
|
chr9_+_106158549
Show fit
|
16.77 |
ENSMUST00000191434.2
|
Poc1a
|
POC1 centriolar protein A
|
|
chr1_-_133681419
Show fit
|
16.54 |
ENSMUST00000125659.8
ENSMUST00000048953.14
ENSMUST00000165602.9
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr15_-_36609812
Show fit
|
16.17 |
ENSMUST00000226496.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1
|
|
chr7_-_126736979
Show fit
|
16.13 |
ENSMUST00000049931.6
|
Spn
|
sialophorin
|
|
chr2_+_118644717
Show fit
|
16.10 |
ENSMUST00000028803.14
ENSMUST00000126045.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding
|
|
chr11_+_87684548
Show fit
|
16.05 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase
|
|
chr6_-_125168453
Show fit
|
15.89 |
ENSMUST00000189959.2
|
Ncapd2
|
non-SMC condensin I complex, subunit D2
|
|
chr9_+_106158212
Show fit
|
15.54 |
ENSMUST00000072206.14
|
Poc1a
|
POC1 centriolar protein A
|
|
chr5_+_123887759
Show fit
|
14.68 |
ENSMUST00000031366.12
|
Kntc1
|
kinetochore associated 1
|
|
chr5_+_33815466
Show fit
|
14.64 |
ENSMUST00000074849.13
ENSMUST00000079534.11
ENSMUST00000201633.2
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr2_+_118644675
Show fit
|
14.41 |
ENSMUST00000110842.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding
|
|
chr2_-_172212426
Show fit
|
14.14 |
ENSMUST00000109139.8
ENSMUST00000028997.8
ENSMUST00000109140.10
|
Aurka
|
aurora kinase A
|
|
chr17_-_80514725
Show fit
|
13.82 |
ENSMUST00000234696.2
ENSMUST00000235069.2
ENSMUST00000063417.11
|
Srsf7
|
serine and arginine-rich splicing factor 7
|
|
chr1_+_135060431
Show fit
|
13.68 |
ENSMUST00000187985.7
ENSMUST00000049449.11
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr7_+_28140352
Show fit
|
13.67 |
ENSMUST00000078845.13
|
Gmfg
|
glia maturation factor, gamma
|
|
chr3_+_146110709
Show fit
|
13.44 |
ENSMUST00000129978.2
|
Ssx2ip
|
synovial sarcoma, X 2 interacting protein
|
|
chr19_-_41790458
Show fit
|
13.18 |
ENSMUST00000026150.15
ENSMUST00000163265.9
ENSMUST00000177495.2
|
Arhgap19
|
Rho GTPase activating protein 19
|
|
chr2_+_118644475
Show fit
|
12.89 |
ENSMUST00000134661.8
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding
|
|
chr14_+_46997984
Show fit
|
12.67 |
ENSMUST00000067426.6
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3
|
|
chr11_+_58531220
Show fit
|
12.40 |
ENSMUST00000075084.5
|
Trim58
|
tripartite motif-containing 58
|
|
chr7_-_16657825
Show fit
|
12.32 |
ENSMUST00000019514.10
|
Calm3
|
calmodulin 3
|
|
chrX_-_101200670
Show fit
|
12.18 |
ENSMUST00000056904.3
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 like
|
|
chr2_-_127673738
Show fit
|
12.02 |
ENSMUST00000028858.8
|
Bub1
|
BUB1, mitotic checkpoint serine/threonine kinase
|
|
chr5_+_33815910
Show fit
|
11.99 |
ENSMUST00000114426.10
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr1_-_131065967
Show fit
|
11.72 |
ENSMUST00000189756.2
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3
|
|
chr3_+_137570334
Show fit
|
11.52 |
ENSMUST00000174561.8
ENSMUST00000173790.8
|
H2az1
|
H2A.Z variant histone 1
|
|
chr1_-_133728779
Show fit
|
11.50 |
ENSMUST00000143567.8
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr3_+_137570248
Show fit
|
11.38 |
ENSMUST00000041045.14
|
H2az1
|
H2A.Z variant histone 1
|
|
chrX_+_55500170
Show fit
|
11.34 |
ENSMUST00000039374.9
ENSMUST00000101553.9
ENSMUST00000186445.7
|
Ints6l
|
integrator complex subunit 6 like
|
|
chr8_+_85696453
Show fit
|
11.34 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2
|
|
chr15_-_97742134
Show fit
|
11.31 |
ENSMUST00000119670.8
ENSMUST00000116409.9
|
Hdac7
|
histone deacetylase 7
|
|
chr8_+_85696396
Show fit
|
11.23 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2
|
|
chr7_+_120450406
Show fit
|
11.12 |
ENSMUST00000143322.9
ENSMUST00000106488.3
|
Eef2k
|
eukaryotic elongation factor-2 kinase
|
|
chr7_+_28140450
Show fit
|
11.11 |
ENSMUST00000135686.2
|
Gmfg
|
glia maturation factor, gamma
|
|
chr1_+_134890288
Show fit
|
10.79 |
ENSMUST00000027687.8
|
Ube2t
|
ubiquitin-conjugating enzyme E2T
|
|
chr1_+_82817170
Show fit
|
10.69 |
ENSMUST00000189220.7
ENSMUST00000113444.8
|
Agfg1
|
ArfGAP with FG repeats 1
|
|
chr1_+_82817388
Show fit
|
10.69 |
ENSMUST00000190052.7
ENSMUST00000063380.11
ENSMUST00000187899.7
ENSMUST00000186302.7
ENSMUST00000190046.7
|
Agfg1
|
ArfGAP with FG repeats 1
|
|
chr4_-_44167509
Show fit
|
10.45 |
ENSMUST00000098098.9
|
Rnf38
|
ring finger protein 38
|
|
chr7_+_12758046
Show fit
|
10.42 |
ENSMUST00000005705.8
|
Trim28
|
tripartite motif-containing 28
|
|
chr2_-_152673585
Show fit
|
10.41 |
ENSMUST00000156688.2
ENSMUST00000007803.12
|
Bcl2l1
|
BCL2-like 1
|
|
chrX_+_133208833
Show fit
|
10.26 |
ENSMUST00000081064.12
ENSMUST00000101251.8
ENSMUST00000129782.2
|
Cenpi
|
centromere protein I
|
|
chr1_+_135060994
Show fit
|
10.26 |
ENSMUST00000167080.3
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr4_-_131802561
Show fit
|
10.12 |
ENSMUST00000105970.8
ENSMUST00000105975.8
|
Epb41
|
erythrocyte membrane protein band 4.1
|
|
chr18_+_34758062
Show fit
|
10.03 |
ENSMUST00000166044.3
|
Kif20a
|
kinesin family member 20A
|
|
chr11_-_120239339
Show fit
|
9.96 |
ENSMUST00000071555.13
|
Actg1
|
actin, gamma, cytoplasmic 1
|
|
chr17_+_27136065
Show fit
|
9.91 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B
|
|
chr2_+_30306116
Show fit
|
9.84 |
ENSMUST00000113601.10
ENSMUST00000113603.10
|
Ptpa
|
protein phosphatase 2 protein activator
|
|
chr7_+_126461601
Show fit
|
9.69 |
ENSMUST00000132808.2
|
Hirip3
|
HIRA interacting protein 3
|
|
chr11_-_120239301
Show fit
|
9.43 |
ENSMUST00000062147.14
ENSMUST00000128055.2
|
Actg1
|
actin, gamma, cytoplasmic 1
|
|
chr2_+_30306045
Show fit
|
9.24 |
ENSMUST00000042055.10
|
Ptpa
|
protein phosphatase 2 protein activator
|
|
chr10_+_45453907
Show fit
|
9.20 |
ENSMUST00000037044.13
|
Hace1
|
HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1
|
|
chr11_-_109364046
Show fit
|
8.93 |
ENSMUST00000070872.13
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6
|
|
chr7_-_45084012
Show fit
|
8.86 |
ENSMUST00000107771.12
ENSMUST00000211666.2
|
Ruvbl2
|
RuvB-like protein 2
|
|
chr7_-_45083688
Show fit
|
8.82 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2
|
|
chr12_-_112792971
Show fit
|
8.80 |
ENSMUST00000062092.7
ENSMUST00000220899.2
|
Cdca4
|
cell division cycle associated 4
|
|
chr11_+_104468107
Show fit
|
8.70 |
ENSMUST00000106956.10
|
Myl4
|
myosin, light polypeptide 4
|
|
chr4_-_131802606
Show fit
|
8.65 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1
|
|
chr17_-_46513499
Show fit
|
8.63 |
ENSMUST00000024749.9
|
Polh
|
polymerase (DNA directed), eta (RAD 30 related)
|
|
chr5_-_8472696
Show fit
|
8.60 |
ENSMUST00000171808.8
|
Dbf4
|
DBF4 zinc finger
|
|
chr7_+_126461117
Show fit
|
8.39 |
ENSMUST00000037248.10
|
Hirip3
|
HIRA interacting protein 3
|
|
chr18_+_34757687
Show fit
|
8.32 |
ENSMUST00000237407.2
|
Kif20a
|
kinesin family member 20A
|
|
chr14_+_46998004
Show fit
|
8.17 |
ENSMUST00000227149.2
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3
|
|
chr1_-_45542442
Show fit
|
8.11 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr4_-_44167904
Show fit
|
8.10 |
ENSMUST00000102934.9
|
Rnf38
|
ring finger protein 38
|
|
chr5_-_8472582
Show fit
|
8.07 |
ENSMUST00000168500.8
ENSMUST00000002368.16
|
Dbf4
|
DBF4 zinc finger
|
|
chr18_+_56840813
Show fit
|
8.06 |
ENSMUST00000025486.9
|
Lmnb1
|
lamin B1
|
|
chr8_-_72175949
Show fit
|
8.05 |
ENSMUST00000125092.2
|
Fcho1
|
FCH domain only 1
|
|
chr15_-_36609208
Show fit
|
7.98 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1
|
|
chr18_+_34757666
Show fit
|
7.93 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A
|
|
chr8_+_84441806
Show fit
|
7.90 |
ENSMUST00000019576.15
|
Ddx39a
|
DEAD box helicase 39a
|
|
chr3_+_116388600
Show fit
|
7.85 |
ENSMUST00000198386.5
ENSMUST00000198311.5
ENSMUST00000197335.2
|
Sass6
|
SAS-6 centriolar assembly protein
|
|
chr8_+_104977493
Show fit
|
7.82 |
ENSMUST00000034342.13
ENSMUST00000212433.2
ENSMUST00000211809.2
|
Cklf
|
chemokine-like factor
|
|
chr15_-_97742045
Show fit
|
7.81 |
ENSMUST00000120683.8
|
Hdac7
|
histone deacetylase 7
|
|
chr4_-_129636073
Show fit
|
7.61 |
ENSMUST00000066257.6
|
Khdrbs1
|
KH domain containing, RNA binding, signal transduction associated 1
|
|
chr11_-_120238917
Show fit
|
7.52 |
ENSMUST00000106215.11
|
Actg1
|
actin, gamma, cytoplasmic 1
|
|
chr8_+_84442133
Show fit
|
7.38 |
ENSMUST00000109810.2
|
Ddx39a
|
DEAD box helicase 39a
|
|
chr2_+_29780532
Show fit
|
7.37 |
ENSMUST00000113764.4
|
Odf2
|
outer dense fiber of sperm tails 2
|
|
chr11_-_85030761
Show fit
|
7.06 |
ENSMUST00000108075.9
|
Usp32
|
ubiquitin specific peptidase 32
|
|
chr1_-_191307648
Show fit
|
7.01 |
ENSMUST00000027933.11
|
Dtl
|
denticleless E3 ubiquitin protein ligase
|
|
chr10_-_75600100
Show fit
|
6.93 |
ENSMUST00000218469.2
ENSMUST00000001712.8
|
Cabin1
|
calcineurin binding protein 1
|
|
chrX_+_74557905
Show fit
|
6.83 |
ENSMUST00000114070.10
ENSMUST00000033540.6
|
Vbp1
|
von Hippel-Lindau binding protein 1
|
|
chr5_+_33978035
Show fit
|
6.74 |
ENSMUST00000075812.11
ENSMUST00000114397.9
ENSMUST00000155880.8
|
Nsd2
|
nuclear receptor binding SET domain protein 2
|
|
chr13_-_55677109
Show fit
|
6.65 |
ENSMUST00000223563.2
|
Dok3
|
docking protein 3
|
|
chr8_+_84335176
Show fit
|
6.63 |
ENSMUST00000212300.2
|
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1
|
|
chr17_-_48716756
Show fit
|
6.58 |
ENSMUST00000160319.8
ENSMUST00000159535.2
ENSMUST00000078800.13
ENSMUST00000046719.14
ENSMUST00000162460.8
|
Nfya
|
nuclear transcription factor-Y alpha
|
|
chr10_+_127126643
Show fit
|
6.55 |
ENSMUST00000026475.15
ENSMUST00000139091.2
ENSMUST00000230446.2
|
Ddit3
Ddit3
|
DNA-damage inducible transcript 3
DNA-damage inducible transcript 3
|
|
chr8_-_73302068
Show fit
|
6.43 |
ENSMUST00000058534.7
|
Med26
|
mediator complex subunit 26
|
|
chr8_+_84441854
Show fit
|
6.39 |
ENSMUST00000172396.8
|
Ddx39a
|
DEAD box helicase 39a
|
|
chr4_-_83404690
Show fit
|
6.36 |
ENSMUST00000107214.9
ENSMUST00000107215.9
ENSMUST00000030207.15
|
Psip1
|
PC4 and SFRS1 interacting protein 1
|
|
chr15_-_79718423
Show fit
|
6.32 |
ENSMUST00000109623.8
ENSMUST00000109625.8
ENSMUST00000023060.13
ENSMUST00000089299.6
|
Cbx6
Npcd
|
chromobox 6
neuronal pentraxin chromo domain
|
|
chr5_-_33809872
Show fit
|
6.31 |
ENSMUST00000057551.14
|
Slbp
|
stem-loop binding protein
|
|
chr11_+_104467791
Show fit
|
6.29 |
ENSMUST00000106957.8
|
Myl4
|
myosin, light polypeptide 4
|
|
chr7_+_46496506
Show fit
|
6.28 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A
|
|
chr5_-_122510292
Show fit
|
6.28 |
ENSMUST00000031419.6
|
Fam216a
|
family with sequence similarity 216, member A
|
|
chr6_-_47571901
Show fit
|
6.14 |
ENSMUST00000081721.13
ENSMUST00000114618.8
ENSMUST00000114616.8
|
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit
|
|
chr14_+_47710005
Show fit
|
6.11 |
ENSMUST00000043112.9
ENSMUST00000163324.8
ENSMUST00000228668.2
ENSMUST00000168833.9
|
Fbxo34
|
F-box protein 34
|
|
chr2_-_164876690
Show fit
|
5.93 |
ENSMUST00000122070.2
ENSMUST00000121377.8
ENSMUST00000153905.2
ENSMUST00000040381.15
|
Ncoa5
|
nuclear receptor coactivator 5
|
|
chr1_-_75482975
Show fit
|
5.87 |
ENSMUST00000113567.10
ENSMUST00000113565.3
|
Obsl1
|
obscurin-like 1
|
|
chr7_+_92524495
Show fit
|
5.85 |
ENSMUST00000207594.2
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C)
|
|
chr5_+_33815892
Show fit
|
5.85 |
ENSMUST00000152847.8
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr13_+_51799268
Show fit
|
5.81 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr7_+_92524460
Show fit
|
5.74 |
ENSMUST00000076052.8
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C)
|
|
chr7_+_46496929
Show fit
|
5.73 |
ENSMUST00000132157.2
ENSMUST00000210631.2
|
Ldha
|
lactate dehydrogenase A
|
|
chr7_-_126461000
Show fit
|
5.70 |
ENSMUST00000106343.3
ENSMUST00000205349.2
ENSMUST00000206349.2
|
Ino80e
|
INO80 complex subunit E
|
|
chr19_-_4861536
Show fit
|
5.68 |
ENSMUST00000179909.8
ENSMUST00000172000.3
ENSMUST00000180008.2
ENSMUST00000113793.4
ENSMUST00000006625.8
|
Gm21992
Rbm14
|
predicted gene 21992
RNA binding motif protein 14
|
|
chr7_+_46496552
Show fit
|
5.60 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A
|
|
chr1_-_75119277
Show fit
|
5.48 |
ENSMUST00000168720.8
ENSMUST00000041213.12
ENSMUST00000189809.2
|
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1
|
|
chr2_+_122479770
Show fit
|
5.47 |
ENSMUST00000047498.15
ENSMUST00000110512.4
|
AA467197
|
expressed sequence AA467197
|
|
chr4_+_132262853
Show fit
|
5.47 |
ENSMUST00000094657.10
ENSMUST00000105940.10
ENSMUST00000105939.10
ENSMUST00000150207.8
|
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8
|
|
chr17_-_47143214
Show fit
|
5.40 |
ENSMUST00000233537.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like
|
|
chr14_+_47710085
Show fit
|
5.36 |
ENSMUST00000095941.9
|
Fbxo34
|
F-box protein 34
|
|
chr10_+_80100812
Show fit
|
5.33 |
ENSMUST00000105362.8
ENSMUST00000105361.10
|
Dazap1
|
DAZ associated protein 1
|
|
chr11_-_33463722
Show fit
|
5.23 |
ENSMUST00000102815.10
|
Ranbp17
|
RAN binding protein 17
|
|
chr11_+_22940519
Show fit
|
5.21 |
ENSMUST00000173867.8
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta)
|
|
chr5_+_76331727
Show fit
|
5.20 |
ENSMUST00000031144.14
|
Tmem165
|
transmembrane protein 165
|
|
chr7_-_126399778
Show fit
|
5.16 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate
|
|
chr10_+_127512933
Show fit
|
5.09 |
ENSMUST00000118612.8
ENSMUST00000048099.5
|
Nemp1
|
nuclear envelope integral membrane protein 1
|
|
chr5_+_137627431
Show fit
|
4.93 |
ENSMUST00000176667.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr8_+_84334805
Show fit
|
4.92 |
ENSMUST00000005620.10
|
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1
|
|
chr11_-_69811347
Show fit
|
4.90 |
ENSMUST00000108610.8
|
Eif5a
|
eukaryotic translation initiation factor 5A
|
|
chr7_+_44465353
Show fit
|
4.86 |
ENSMUST00000208626.2
ENSMUST00000057195.17
|
Nup62
|
nucleoporin 62
|
|
chr7_-_126101484
Show fit
|
4.81 |
ENSMUST00000166682.9
|
Atxn2l
|
ataxin 2-like
|
|
chr6_+_117883732
Show fit
|
4.71 |
ENSMUST00000179224.8
ENSMUST00000035493.14
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr11_-_69649004
Show fit
|
4.70 |
ENSMUST00000071213.4
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A
|
|
chr11_+_22940599
Show fit
|
4.66 |
ENSMUST00000020562.5
|
Cct4
|
chaperonin containing Tcp1, subunit 4 (delta)
|
|
chr17_+_46513666
Show fit
|
4.62 |
ENSMUST00000087031.7
|
Xpo5
|
exportin 5
|
|
chr4_-_11007635
Show fit
|
4.59 |
ENSMUST00000054776.4
|
Plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2
|
|
chr2_-_153286361
Show fit
|
4.57 |
ENSMUST00000109784.2
|
Nol4l
|
nucleolar protein 4-like
|
|
chr4_-_44168339
Show fit
|
4.50 |
ENSMUST00000045793.15
|
Rnf38
|
ring finger protein 38
|
|
chr6_+_117883783
Show fit
|
4.47 |
ENSMUST00000177918.8
ENSMUST00000163168.9
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr7_-_126461522
Show fit
|
4.38 |
ENSMUST00000206968.2
|
Ino80e
|
INO80 complex subunit E
|
|
chr17_-_35383867
Show fit
|
4.32 |
ENSMUST00000025253.12
|
Prrc2a
|
proline-rich coiled-coil 2A
|
|
chr10_+_80100868
Show fit
|
4.19 |
ENSMUST00000092305.6
|
Dazap1
|
DAZ associated protein 1
|
|
chrX_+_149829131
Show fit
|
4.17 |
ENSMUST00000112685.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr3_+_103821413
Show fit
|
4.17 |
ENSMUST00000051139.13
ENSMUST00000068879.11
|
Rsbn1
|
rosbin, round spermatid basic protein 1
|
|
chr17_+_28059129
Show fit
|
4.17 |
ENSMUST00000233657.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C
|
|
chr3_+_30910089
Show fit
|
4.16 |
ENSMUST00000108261.8
ENSMUST00000108259.8
ENSMUST00000166278.7
ENSMUST00000046748.13
ENSMUST00000194979.6
|
Gpr160
|
G protein-coupled receptor 160
|
|
chr2_+_4564553
Show fit
|
4.15 |
ENSMUST00000176828.8
|
Frmd4a
|
FERM domain containing 4A
|
|
chr7_+_44465806
Show fit
|
4.14 |
ENSMUST00000207103.2
ENSMUST00000118125.9
|
Nup62
Il4i1
|
nucleoporin 62
interleukin 4 induced 1
|
|
chr11_-_69649452
Show fit
|
4.14 |
ENSMUST00000058470.16
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A
|
|
chr8_+_121264161
Show fit
|
4.05 |
ENSMUST00000118136.2
|
Gse1
|
genetic suppressor element 1, coiled-coil protein
|
|
chr2_-_103627937
Show fit
|
4.05 |
ENSMUST00000028607.13
|
Caprin1
|
cell cycle associated protein 1
|
|
chr7_-_126101555
Show fit
|
4.04 |
ENSMUST00000167759.8
|
Atxn2l
|
ataxin 2-like
|
|
chr11_+_116324913
Show fit
|
4.04 |
ENSMUST00000057676.7
|
Ubald2
|
UBA-like domain containing 2
|
|
chr18_+_61058684
Show fit
|
4.03 |
ENSMUST00000102888.10
ENSMUST00000025519.11
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr17_-_27247891
Show fit
|
3.96 |
ENSMUST00000078691.12
|
Bak1
|
BCL2-antagonist/killer 1
|
|
chr5_-_31377847
Show fit
|
3.96 |
ENSMUST00000202294.4
ENSMUST00000031032.11
|
Ppm1g
|
protein phosphatase 1G (formerly 2C), magnesium-dependent, gamma isoform
|
|
chr2_+_30697838
Show fit
|
3.94 |
ENSMUST00000041830.10
ENSMUST00000152374.8
|
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1
|
|
chr7_-_126101245
Show fit
|
3.90 |
ENSMUST00000179818.3
|
Atxn2l
|
ataxin 2-like
|
|
chr17_-_80369626
Show fit
|
3.87 |
ENSMUST00000184635.8
|
Hnrnpll
|
heterogeneous nuclear ribonucleoprotein L-like
|
|
chr7_-_16121716
Show fit
|
3.86 |
ENSMUST00000211741.2
ENSMUST00000210999.2
|
Sae1
|
SUMO1 activating enzyme subunit 1
|
|
chr11_-_116743898
Show fit
|
3.83 |
ENSMUST00000190993.7
ENSMUST00000092404.13
|
Srsf2
|
serine and arginine-rich splicing factor 2
|
|
chr15_-_103148239
Show fit
|
3.82 |
ENSMUST00000118152.8
|
Cbx5
|
chromobox 5
|
|
chrX_+_20554193
Show fit
|
3.81 |
ENSMUST00000115364.8
|
Cdk16
|
cyclin-dependent kinase 16
|
|
chr7_+_118311740
Show fit
|
3.79 |
ENSMUST00000106557.8
|
Ccp110
|
centriolar coiled coil protein 110
|
|
chr10_-_128401773
Show fit
|
3.78 |
ENSMUST00000026425.13
ENSMUST00000131728.4
|
Pa2g4
|
proliferation-associated 2G4
|
|
chr15_-_34443054
Show fit
|
3.75 |
ENSMUST00000142643.2
|
Rpl30
|
ribosomal protein L30
|
|
chr17_+_28059099
Show fit
|
3.74 |
ENSMUST00000233752.2
|
Snrpc
|
U1 small nuclear ribonucleoprotein C
|
|
chr2_-_25162347
Show fit
|
3.74 |
ENSMUST00000028342.7
|
Ssna1
|
SS nuclear autoantigen 1
|
|
chr3_-_51248032
Show fit
|
3.67 |
ENSMUST00000062009.14
ENSMUST00000194641.6
|
Elf2
|
E74-like factor 2
|
|
chrX_+_20554618
Show fit
|
3.67 |
ENSMUST00000033380.7
|
Cdk16
|
cyclin-dependent kinase 16
|
|
chr11_+_23615612
Show fit
|
3.63 |
ENSMUST00000109525.8
ENSMUST00000020520.11
|
Pus10
|
pseudouridylate synthase 10
|
|
chr11_-_69811717
Show fit
|
3.60 |
ENSMUST00000152589.2
ENSMUST00000108612.8
ENSMUST00000108611.8
|
Eif5a
|
eukaryotic translation initiation factor 5A
|
|
chr3_-_108819003
Show fit
|
3.59 |
ENSMUST00000029480.9
|
Prpf38b
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B
|
|
chr11_-_74614654
Show fit
|
3.59 |
ENSMUST00000102520.9
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1
|
|
chr2_-_5900130
Show fit
|
3.53 |
ENSMUST00000026926.5
ENSMUST00000193792.6
ENSMUST00000102981.10
|
Sec61a2
|
Sec61, alpha subunit 2 (S. cerevisiae)
|
|
chr2_-_103627334
Show fit
|
3.52 |
ENSMUST00000111147.8
|
Caprin1
|
cell cycle associated protein 1
|
|
chr2_-_74409225
Show fit
|
3.49 |
ENSMUST00000134168.2
ENSMUST00000111993.9
ENSMUST00000064503.13
|
Lnpk
|
lunapark, ER junction formation factor
|
|
chr5_-_137529251
Show fit
|
3.46 |
ENSMUST00000132525.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2
|
|
chr7_+_44465714
Show fit
|
3.45 |
ENSMUST00000208172.2
|
Nup62
|
nucleoporin 62
|
|
chr17_+_28059036
Show fit
|
3.43 |
ENSMUST00000071006.9
|
Snrpc
|
U1 small nuclear ribonucleoprotein C
|
|
chr8_-_106052884
Show fit
|
3.40 |
ENSMUST00000210412.2
ENSMUST00000210801.2
ENSMUST00000070508.8
|
Lrrc29
|
leucine rich repeat containing 29
|
|
chr18_-_34757653
Show fit
|
3.37 |
ENSMUST00000003876.10
ENSMUST00000115766.8
ENSMUST00000097626.10
ENSMUST00000115765.2
|
Brd8
|
bromodomain containing 8
|
|
chr4_-_44168252
Show fit
|
3.34 |
ENSMUST00000145760.8
ENSMUST00000128426.8
|
Rnf38
|
ring finger protein 38
|
|
chr14_+_8508484
Show fit
|
3.32 |
ENSMUST00000065865.10
ENSMUST00000225891.2
|
Thoc7
|
THO complex 7
|
|
chr1_-_161704224
Show fit
|
3.29 |
ENSMUST00000048377.11
|
Suco
|
SUN domain containing ossification factor
|
|
chr10_-_22607817
Show fit
|
3.29 |
ENSMUST00000095794.4
|
Tbpl1
|
TATA box binding protein-like 1
|
|
chr4_+_149569672
Show fit
|
3.27 |
ENSMUST00000124413.8
ENSMUST00000141293.8
|
Lzic
|
leucine zipper and CTNNBIP1 domain containing
|
|
chr3_+_88204454
Show fit
|
3.26 |
ENSMUST00000164166.8
ENSMUST00000168062.8
|
Cct3
|
chaperonin containing Tcp1, subunit 3 (gamma)
|
|
chr15_-_75941530
Show fit
|
3.25 |
ENSMUST00000002603.12
ENSMUST00000063747.12
ENSMUST00000109946.9
|
Scrib
|
scribbled planar cell polarity
|
|
chr7_-_101899294
Show fit
|
3.24 |
ENSMUST00000106923.2
ENSMUST00000098230.11
|
Rhog
|
ras homolog family member G
|
|
chr4_-_148711211
Show fit
|
3.21 |
ENSMUST00000186947.7
ENSMUST00000105699.8
|
Tardbp
|
TAR DNA binding protein
|
|
chr6_-_72416531
Show fit
|
3.21 |
ENSMUST00000205335.2
ENSMUST00000206692.2
ENSMUST00000059472.10
|
Mat2a
|
methionine adenosyltransferase II, alpha
|
|
chr6_-_28261881
Show fit
|
3.19 |
ENSMUST00000115320.8
ENSMUST00000123098.8
ENSMUST00000115321.9
ENSMUST00000155494.2
|
Zfp800
|
zinc finger protein 800
|
|
chr6_+_119456629
Show fit
|
3.17 |
ENSMUST00000032094.7
|
Fbxl14
|
F-box and leucine-rich repeat protein 14
|
|
chr2_-_34803988
Show fit
|
3.15 |
ENSMUST00000028232.7
ENSMUST00000202907.2
|
Phf19
|
PHD finger protein 19
|