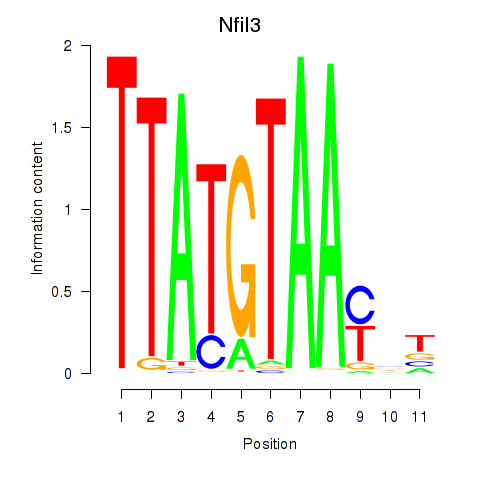
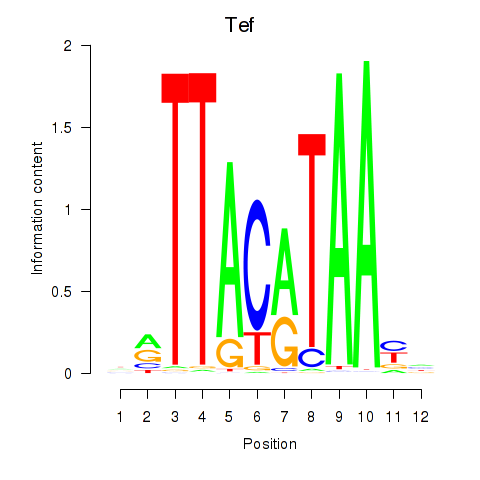
Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nfil3_Tef
Z-value: 2.73
Transcription factors associated with Nfil3_Tef
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfil3
|
ENSMUSG00000056749.8 | nuclear factor, interleukin 3, regulated |
|
Tef
|
ENSMUSG00000022389.16 | thyrotroph embryonic factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfil3 | mm39_v1_chr13_-_53135064_53135114 | 0.70 | 2.2e-06 | Click! |
| Tef | mm39_v1_chr15_+_81686622_81686779 | -0.01 | 9.6e-01 | Click! |
Activity profile of Nfil3_Tef motif
Sorted Z-values of Nfil3_Tef motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_26534730 | 39.68 |
ENSMUST00000005685.15
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr14_+_66208498 | 36.97 |
ENSMUST00000128539.8
|
Clu
|
clusterin |
| chr14_+_66208613 | 32.96 |
ENSMUST00000144619.2
|
Clu
|
clusterin |
| chr14_+_66208253 | 32.82 |
ENSMUST00000138191.8
|
Clu
|
clusterin |
| chr14_+_66208059 | 30.61 |
ENSMUST00000127387.8
|
Clu
|
clusterin |
| chr8_-_93924426 | 25.25 |
ENSMUST00000034172.8
|
Ces1d
|
carboxylesterase 1D |
| chr16_+_22769822 | 23.58 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr16_+_22769844 | 23.02 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr5_-_92475927 | 22.64 |
ENSMUST00000113093.5
|
Cxcl9
|
chemokine (C-X-C motif) ligand 9 |
| chr7_+_26006594 | 22.05 |
ENSMUST00000098657.5
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr8_-_93857899 | 21.99 |
ENSMUST00000034189.17
|
Ces1c
|
carboxylesterase 1C |
| chr4_-_60538151 | 21.76 |
ENSMUST00000098047.3
|
Mup10
|
major urinary protein 10 |
| chr14_+_40827317 | 16.39 |
ENSMUST00000047286.7
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr8_-_93806593 | 16.16 |
ENSMUST00000109582.3
|
Ces1b
|
carboxylesterase 1B |
| chr9_-_78254422 | 16.12 |
ENSMUST00000034902.12
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr2_+_102536701 | 15.92 |
ENSMUST00000123759.8
ENSMUST00000005220.11 ENSMUST00000111212.8 |
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr3_-_113368407 | 15.42 |
ENSMUST00000106540.8
|
Amy1
|
amylase 1, salivary |
| chr5_-_147831610 | 14.99 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr14_+_40827108 | 14.69 |
ENSMUST00000224514.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr3_-_129126362 | 14.18 |
ENSMUST00000029658.14
|
Enpep
|
glutamyl aminopeptidase |
| chr7_+_119125443 | 14.13 |
ENSMUST00000207440.2
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr14_+_40826970 | 14.05 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr7_-_81356653 | 13.44 |
ENSMUST00000026922.15
|
Homer2
|
homer scaffolding protein 2 |
| chr7_+_119125546 | 13.28 |
ENSMUST00000207387.2
ENSMUST00000207813.2 |
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr7_-_81356557 | 13.18 |
ENSMUST00000207983.2
|
Homer2
|
homer scaffolding protein 2 |
| chr9_-_78254443 | 13.17 |
ENSMUST00000129247.2
|
Gsta2
|
glutathione S-transferase, alpha 2 (Yc2) |
| chr12_-_81014849 | 13.16 |
ENSMUST00000095572.5
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr7_+_119125426 | 12.88 |
ENSMUST00000066465.3
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr12_-_81014755 | 12.33 |
ENSMUST00000218342.2
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr14_+_4230569 | 12.17 |
ENSMUST00000090543.6
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr6_+_133082202 | 11.32 |
ENSMUST00000191462.2
|
Smim10l1
|
small integral membrane protein 10 like 1 |
| chr10_-_31321793 | 11.22 |
ENSMUST00000213639.2
ENSMUST00000215515.2 ENSMUST00000214644.2 ENSMUST00000213528.2 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr5_+_90608751 | 10.95 |
ENSMUST00000031314.10
|
Alb
|
albumin |
| chr3_+_63203516 | 10.44 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chr15_-_76191301 | 10.42 |
ENSMUST00000171340.9
ENSMUST00000023222.13 ENSMUST00000164189.2 |
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr15_+_7159038 | 10.41 |
ENSMUST00000067190.12
ENSMUST00000164529.9 |
Lifr
|
LIF receptor alpha |
| chr19_+_31846154 | 10.38 |
ENSMUST00000224564.2
ENSMUST00000224304.2 ENSMUST00000075838.8 ENSMUST00000224400.2 |
A1cf
|
APOBEC1 complementation factor |
| chr19_+_23118545 | 9.91 |
ENSMUST00000036884.3
|
Klf9
|
Kruppel-like factor 9 |
| chr6_-_119444157 | 9.78 |
ENSMUST00000118120.8
|
Wnt5b
|
wingless-type MMTV integration site family, member 5B |
| chr11_-_43792013 | 9.70 |
ENSMUST00000067258.9
ENSMUST00000139906.2 |
Adra1b
|
adrenergic receptor, alpha 1b |
| chr10_-_78300802 | 9.62 |
ENSMUST00000041616.15
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr4_-_104733580 | 9.45 |
ENSMUST00000064873.9
ENSMUST00000106808.10 ENSMUST00000048947.15 |
C8a
|
complement component 8, alpha polypeptide |
| chr2_+_129642371 | 9.18 |
ENSMUST00000165413.9
ENSMUST00000166282.3 |
Stk35
|
serine/threonine kinase 35 |
| chr9_+_78137927 | 8.90 |
ENSMUST00000098537.4
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya) |
| chr1_-_140111138 | 8.87 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr1_-_140111018 | 8.50 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr4_+_47208004 | 8.44 |
ENSMUST00000082303.13
ENSMUST00000102917.11 |
Col15a1
|
collagen, type XV, alpha 1 |
| chr5_-_89605622 | 8.16 |
ENSMUST00000049209.13
|
Gc
|
vitamin D binding protein |
| chr19_-_38113696 | 8.02 |
ENSMUST00000025951.14
ENSMUST00000237287.2 |
Rbp4
|
retinol binding protein 4, plasma |
| chr11_+_94219046 | 8.00 |
ENSMUST00000021227.6
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr16_+_3702523 | 7.90 |
ENSMUST00000176625.8
ENSMUST00000186375.8 |
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr14_-_45626237 | 7.85 |
ENSMUST00000227865.2
ENSMUST00000226856.2 ENSMUST00000226276.2 ENSMUST00000046191.9 |
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr16_+_3702604 | 7.84 |
ENSMUST00000115860.8
|
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr2_+_103858066 | 7.78 |
ENSMUST00000028603.10
|
Fbxo3
|
F-box protein 3 |
| chr15_-_77283286 | 7.76 |
ENSMUST00000175919.8
ENSMUST00000176074.9 |
Apol7a
|
apolipoprotein L 7a |
| chr8_-_72966840 | 7.67 |
ENSMUST00000238973.2
|
Cib3
|
calcium and integrin binding family member 3 |
| chr11_-_106107132 | 7.53 |
ENSMUST00000002043.10
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr5_-_130053120 | 7.47 |
ENSMUST00000161640.8
ENSMUST00000161884.2 ENSMUST00000161094.8 |
Asl
|
argininosuccinate lyase |
| chr11_+_115353290 | 7.44 |
ENSMUST00000106532.4
ENSMUST00000092445.12 ENSMUST00000153466.2 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr9_+_78164402 | 7.41 |
ENSMUST00000217203.2
|
Gm3776
|
predicted gene 3776 |
| chr15_+_68800261 | 7.35 |
ENSMUST00000022954.7
|
Khdrbs3
|
KH domain containing, RNA binding, signal transduction associated 3 |
| chr8_-_123884968 | 7.02 |
ENSMUST00000137998.2
|
Sult5a1
|
sulfotransferase family 5A, member 1 |
| chr17_+_37253802 | 6.53 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr11_+_83594817 | 6.51 |
ENSMUST00000092836.6
|
Wfdc17
|
WAP four-disulfide core domain 17 |
| chr16_+_3702493 | 6.50 |
ENSMUST00000176233.2
|
Gm20695
|
predicted gene 20695 |
| chr9_+_78197205 | 6.47 |
ENSMUST00000119823.8
ENSMUST00000121273.2 |
Gsta5
|
glutathione S-transferase alpha 5 |
| chr15_-_37007623 | 6.37 |
ENSMUST00000078976.9
|
Zfp706
|
zinc finger protein 706 |
| chr19_+_32597379 | 6.33 |
ENSMUST00000236290.2
ENSMUST00000025833.7 |
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr3_+_96152813 | 6.19 |
ENSMUST00000078756.7
ENSMUST00000090779.4 |
H2ac18
Gm20634
|
H2A clustered histone 18 predicted gene 20634 |
| chr8_-_91860576 | 6.15 |
ENSMUST00000120213.9
ENSMUST00000109609.9 |
Aktip
|
AKT interacting protein |
| chr8_-_123885007 | 6.14 |
ENSMUST00000000755.15
|
Sult5a1
|
sulfotransferase family 5A, member 1 |
| chr3_-_96147592 | 6.01 |
ENSMUST00000074976.8
|
H2ac19
|
H2A clustered histone 19 |
| chr6_+_17463748 | 6.01 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr19_-_7780025 | 6.00 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr19_-_7779943 | 5.84 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr8_-_91860655 | 5.78 |
ENSMUST00000125257.3
|
Aktip
|
AKT interacting protein |
| chr14_+_4230658 | 5.70 |
ENSMUST00000225491.2
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr11_+_94218810 | 5.59 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr4_+_43493344 | 5.59 |
ENSMUST00000030181.12
ENSMUST00000107922.3 |
Ccdc107
|
coiled-coil domain containing 107 |
| chr2_+_103858168 | 5.49 |
ENSMUST00000111135.8
ENSMUST00000111136.8 ENSMUST00000102565.4 |
Fbxo3
|
F-box protein 3 |
| chr6_+_124470053 | 5.47 |
ENSMUST00000049124.10
|
C1rl
|
complement component 1, r subcomponent-like |
| chr14_+_48358267 | 5.40 |
ENSMUST00000073150.6
|
Peli2
|
pellino 2 |
| chr3_-_98660781 | 5.37 |
ENSMUST00000094050.11
ENSMUST00000090743.13 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr11_-_69576363 | 5.35 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr3_+_30847024 | 5.33 |
ENSMUST00000029256.9
|
Sec62
|
SEC62 homolog (S. cerevisiae) |
| chr13_-_53135064 | 5.15 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr7_+_112026712 | 5.03 |
ENSMUST00000106643.8
ENSMUST00000033030.14 |
Parva
|
parvin, alpha |
| chr19_-_11058452 | 4.77 |
ENSMUST00000025636.8
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr14_+_79086492 | 4.74 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr11_+_60619224 | 4.73 |
ENSMUST00000018743.5
|
Mief2
|
mitochondrial elongation factor 2 |
| chr2_-_37593287 | 4.68 |
ENSMUST00000072186.12
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr3_+_59939175 | 4.62 |
ENSMUST00000029325.5
|
Aadac
|
arylacetamide deacetylase |
| chr17_-_74257164 | 4.55 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr10_-_31321938 | 4.52 |
ENSMUST00000000305.7
|
Tpd52l1
|
tumor protein D52-like 1 |
| chr16_-_11727262 | 4.49 |
ENSMUST00000127972.8
ENSMUST00000121750.2 ENSMUST00000096272.11 ENSMUST00000073371.7 |
Cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr2_-_173060647 | 4.47 |
ENSMUST00000109116.3
ENSMUST00000029018.14 |
Zbp1
|
Z-DNA binding protein 1 |
| chr17_+_44112679 | 4.43 |
ENSMUST00000229744.2
|
Rcan2
|
regulator of calcineurin 2 |
| chr1_-_58625350 | 4.43 |
ENSMUST00000038372.14
ENSMUST00000097724.10 |
Fam126b
|
family with sequence similarity 126, member B |
| chr8_+_46944000 | 4.40 |
ENSMUST00000110372.9
ENSMUST00000130563.2 |
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr4_-_134580604 | 4.38 |
ENSMUST00000030628.15
|
Maco1
|
macoilin 1 |
| chr9_-_71070506 | 4.34 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr13_+_25127127 | 4.13 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr1_+_172525613 | 4.08 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr2_-_60114684 | 4.06 |
ENSMUST00000028356.9
ENSMUST00000074606.11 |
Cd302
|
CD302 antigen |
| chr9_+_78099229 | 4.04 |
ENSMUST00000034903.7
|
Gsta4
|
glutathione S-transferase, alpha 4 |
| chr10_-_116732813 | 3.82 |
ENSMUST00000048229.9
|
Myrfl
|
myelin regulatory factor-like |
| chr5_-_5564730 | 3.70 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr4_+_19575128 | 3.66 |
ENSMUST00000108253.8
ENSMUST00000029888.4 |
Rmdn1
|
regulator of microtubule dynamics 1 |
| chr5_-_5564873 | 3.65 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr2_+_118692435 | 3.63 |
ENSMUST00000028807.6
|
Ivd
|
isovaleryl coenzyme A dehydrogenase |
| chr10_+_23727325 | 3.63 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr7_+_143729250 | 3.63 |
ENSMUST00000105900.9
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr18_+_13139992 | 3.61 |
ENSMUST00000041676.3
ENSMUST00000234084.2 ENSMUST00000234565.2 |
Hrh4
|
histamine receptor H4 |
| chr11_-_78313043 | 3.57 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr17_+_26934617 | 3.55 |
ENSMUST00000062519.14
ENSMUST00000144221.2 ENSMUST00000142539.8 ENSMUST00000151681.2 |
Crebrf
|
CREB3 regulatory factor |
| chr1_-_164281344 | 3.48 |
ENSMUST00000193367.2
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr1_-_92107971 | 3.45 |
ENSMUST00000186002.3
ENSMUST00000097644.9 |
Hdac4
|
histone deacetylase 4 |
| chrX_+_10583629 | 3.41 |
ENSMUST00000115524.8
ENSMUST00000008179.7 ENSMUST00000156321.2 |
Mid1ip1
|
Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) |
| chr1_-_14380418 | 3.37 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr2_-_25359752 | 3.33 |
ENSMUST00000114259.3
ENSMUST00000015234.13 |
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr12_+_21366386 | 3.26 |
ENSMUST00000076813.8
ENSMUST00000221693.2 ENSMUST00000223345.2 ENSMUST00000222344.2 |
Iah1
|
isoamyl acetate-hydrolyzing esterase 1 homolog |
| chr7_+_126380655 | 3.25 |
ENSMUST00000172352.8
ENSMUST00000094037.5 |
Tbx6
|
T-box 6 |
| chr7_+_26508305 | 3.23 |
ENSMUST00000040944.9
|
Cyp2g1
|
cytochrome P450, family 2, subfamily g, polypeptide 1 |
| chr5_+_135038006 | 3.21 |
ENSMUST00000111216.8
ENSMUST00000046999.12 |
Abhd11
|
abhydrolase domain containing 11 |
| chr19_-_34724689 | 3.14 |
ENSMUST00000009522.4
|
Slc16a12
|
solute carrier family 16 (monocarboxylic acid transporters), member 12 |
| chr10_-_125144678 | 3.13 |
ENSMUST00000105257.4
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chr1_-_14380327 | 3.11 |
ENSMUST00000080664.14
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr14_-_73613385 | 3.09 |
ENSMUST00000227454.2
|
Itm2b
|
integral membrane protein 2B |
| chr12_+_37292029 | 3.07 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chr9_+_32305259 | 2.99 |
ENSMUST00000172015.3
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr5_-_52827015 | 2.94 |
ENSMUST00000031069.13
|
Sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr2_-_90735171 | 2.92 |
ENSMUST00000005647.4
|
Ndufs3
|
NADH:ubiquinone oxidoreductase core subunit S3 |
| chr15_+_44482667 | 2.91 |
ENSMUST00000228648.2
ENSMUST00000226165.2 |
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr17_+_8502594 | 2.83 |
ENSMUST00000155364.8
ENSMUST00000046754.15 |
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr13_+_51254852 | 2.81 |
ENSMUST00000095797.6
|
Spin1
|
spindlin 1 |
| chr19_-_42420216 | 2.80 |
ENSMUST00000048630.8
ENSMUST00000238290.2 |
Crtac1
|
cartilage acidic protein 1 |
| chr4_+_101276893 | 2.77 |
ENSMUST00000131397.2
ENSMUST00000133055.8 |
Ak4
|
adenylate kinase 4 |
| chr15_+_44482944 | 2.72 |
ENSMUST00000022964.9
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr3_+_89366425 | 2.70 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr8_+_22550727 | 2.64 |
ENSMUST00000072572.13
ENSMUST00000110737.3 ENSMUST00000131624.2 |
Alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr12_+_86999366 | 2.59 |
ENSMUST00000191463.2
|
Cipc
|
CLOCK interacting protein, circadian |
| chr3_+_129326285 | 2.58 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr12_+_37291632 | 2.58 |
ENSMUST00000049874.14
|
Agmo
|
alkylglycerol monooxygenase |
| chr12_+_37291728 | 2.40 |
ENSMUST00000160768.8
|
Agmo
|
alkylglycerol monooxygenase |
| chr4_+_40269563 | 2.39 |
ENSMUST00000129758.3
|
Smim27
|
small integral membrane protein 27 |
| chr13_+_112600604 | 2.39 |
ENSMUST00000183663.8
ENSMUST00000184311.8 ENSMUST00000183886.8 |
Il6st
|
interleukin 6 signal transducer |
| chr7_+_28937859 | 2.36 |
ENSMUST00000108237.2
|
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr5_+_135038267 | 2.35 |
ENSMUST00000201890.2
ENSMUST00000154469.8 |
Abhd11
|
abhydrolase domain containing 11 |
| chr2_-_12424212 | 2.34 |
ENSMUST00000124603.8
ENSMUST00000129993.3 ENSMUST00000028105.13 |
Mindy3
|
MINDY lysine 48 deubiquitinase 3 |
| chr3_+_138148846 | 2.31 |
ENSMUST00000005964.7
|
Adh5
|
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chr17_-_30845845 | 2.30 |
ENSMUST00000235547.2
ENSMUST00000188852.2 |
Glo1
1700097N02Rik
|
glyoxalase 1 RIKEN cDNA 1700097N02 gene |
| chr19_+_34560922 | 2.27 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr11_-_101676076 | 2.26 |
ENSMUST00000164750.8
ENSMUST00000107176.8 ENSMUST00000017868.7 |
Etv4
|
ets variant 4 |
| chr13_-_67599702 | 2.21 |
ENSMUST00000225479.2
ENSMUST00000057241.15 ENSMUST00000075255.6 |
Zfp874a
|
zinc finger protein 874a |
| chr2_-_136727599 | 2.15 |
ENSMUST00000231720.2
|
ENSMUSG00000116563.2
|
novel protein |
| chr4_-_150087587 | 2.13 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr14_-_45626198 | 2.12 |
ENSMUST00000226590.2
|
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr17_+_14087827 | 2.11 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr17_+_37253916 | 2.10 |
ENSMUST00000173072.2
|
Rnf39
|
ring finger protein 39 |
| chr2_-_25360043 | 2.10 |
ENSMUST00000114251.8
|
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr11_+_83599841 | 2.05 |
ENSMUST00000001009.14
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr12_+_87247297 | 2.04 |
ENSMUST00000182869.2
|
Samd15
|
sterile alpha motif domain containing 15 |
| chr16_-_22258320 | 2.04 |
ENSMUST00000170393.2
|
Etv5
|
ets variant 5 |
| chr10_+_20046748 | 2.02 |
ENSMUST00000215924.2
|
Map7
|
microtubule-associated protein 7 |
| chr10_-_23985432 | 2.02 |
ENSMUST00000041180.7
|
Taar9
|
trace amine-associated receptor 9 |
| chr4_+_48279794 | 2.01 |
ENSMUST00000030029.10
|
Invs
|
inversin |
| chr11_+_21041291 | 2.01 |
ENSMUST00000093290.12
|
Peli1
|
pellino 1 |
| chr6_+_121160626 | 1.99 |
ENSMUST00000118234.8
ENSMUST00000088561.10 ENSMUST00000137432.8 ENSMUST00000120066.8 |
Pex26
|
peroxisomal biogenesis factor 26 |
| chr11_-_73217298 | 1.95 |
ENSMUST00000155630.9
|
Aspa
|
aspartoacylase |
| chr5_-_30401416 | 1.92 |
ENSMUST00000125367.4
|
Adgrf3
|
adhesion G protein-coupled receptor F3 |
| chr14_+_74973081 | 1.90 |
ENSMUST00000177283.8
|
Esd
|
esterase D/formylglutathione hydrolase |
| chr4_-_41774097 | 1.86 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
| chr4_+_55350043 | 1.83 |
ENSMUST00000030134.9
|
Rad23b
|
RAD23 homolog B, nucleotide excision repair protein |
| chr15_-_33687986 | 1.80 |
ENSMUST00000042021.5
|
Tspyl5
|
testis-specific protein, Y-encoded-like 5 |
| chr1_-_84816379 | 1.80 |
ENSMUST00000187818.2
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr15_-_103473481 | 1.79 |
ENSMUST00000228060.2
ENSMUST00000228895.2 ENSMUST00000023134.5 |
Glycam1
|
glycosylation dependent cell adhesion molecule 1 |
| chr15_-_93234681 | 1.78 |
ENSMUST00000080299.7
|
Yaf2
|
YY1 associated factor 2 |
| chr6_+_17463819 | 1.75 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr1_-_14380032 | 1.75 |
ENSMUST00000187790.2
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr11_-_6394385 | 1.72 |
ENSMUST00000109737.9
|
H2az2
|
H2A.Z histone variant 2 |
| chr1_-_160862364 | 1.63 |
ENSMUST00000177003.2
ENSMUST00000159250.9 ENSMUST00000162226.9 |
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr8_+_22682816 | 1.61 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chr15_+_41694317 | 1.55 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr5_-_88823989 | 1.55 |
ENSMUST00000078945.12
|
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr7_+_28937898 | 1.52 |
ENSMUST00000138128.3
ENSMUST00000142519.3 |
Yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr4_+_85123654 | 1.50 |
ENSMUST00000030212.15
ENSMUST00000107189.8 ENSMUST00000107184.8 |
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr7_-_126992776 | 1.50 |
ENSMUST00000165495.2
ENSMUST00000106303.3 ENSMUST00000074249.7 |
E430018J23Rik
|
RIKEN cDNA E430018J23 gene |
| chr5_-_129916283 | 1.46 |
ENSMUST00000094280.4
|
Chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr15_-_38079089 | 1.46 |
ENSMUST00000110336.4
|
Ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr8_+_71849497 | 1.42 |
ENSMUST00000002473.10
|
Babam1
|
BRISC and BRCA1 A complex member 1 |
| chr8_-_34614187 | 1.42 |
ENSMUST00000033910.9
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr8_-_41494890 | 1.38 |
ENSMUST00000051379.14
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr16_-_16345197 | 1.37 |
ENSMUST00000069284.14
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chrX_-_74426221 | 1.36 |
ENSMUST00000147349.8
|
F8
|
coagulation factor VIII |
| chr1_+_11063678 | 1.36 |
ENSMUST00000027056.12
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr8_+_26298502 | 1.34 |
ENSMUST00000033979.6
|
Star
|
steroidogenic acute regulatory protein |
| chr4_+_85123358 | 1.31 |
ENSMUST00000107188.10
|
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr14_+_48358331 | 1.30 |
ENSMUST00000226513.2
|
Peli2
|
pellino 2 |
| chr4_-_119395966 | 1.28 |
ENSMUST00000079611.13
|
Frg2f1
|
FSHD region gene 2 family member 1 |
| chr2_+_85805557 | 1.25 |
ENSMUST00000082191.5
|
Olfr1029
|
olfactory receptor 1029 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfil3_Tef
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.2 | 133.4 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 15.5 | 46.6 | GO:0097037 | heme export(GO:0097037) |
| 15.0 | 45.1 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 8.4 | 25.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 3.7 | 11.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 3.2 | 9.7 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 3.2 | 9.6 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 3.1 | 102.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 2.6 | 10.4 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 2.5 | 7.5 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 2.5 | 17.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 2.4 | 21.5 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 2.0 | 14.2 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 2.0 | 10.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 1.8 | 12.8 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 1.8 | 5.4 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 1.7 | 15.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 1.6 | 11.4 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 1.6 | 8.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 1.6 | 6.3 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.5 | 10.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 1.4 | 4.2 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 1.2 | 3.5 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 1.1 | 26.6 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 1.1 | 7.8 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.1 | 4.3 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.0 | 3.1 | GO:0015881 | creatine transport(GO:0015881) |
| 0.9 | 2.8 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.9 | 4.5 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 0.9 | 5.4 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.9 | 25.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.8 | 22.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.8 | 8.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.8 | 11.8 | GO:0015747 | urate transport(GO:0015747) |
| 0.8 | 3.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.7 | 8.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.7 | 2.7 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.6 | 9.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.6 | 8.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.6 | 1.8 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.6 | 4.8 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.6 | 3.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.6 | 3.4 | GO:0014854 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.5 | 7.5 | GO:0006983 | ER overload response(GO:0006983) |
| 0.5 | 17.9 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.5 | 4.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.5 | 1.9 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.5 | 3.3 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.4 | 3.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.4 | 3.0 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.4 | 6.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.4 | 2.0 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.4 | 2.8 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.4 | 39.8 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.4 | 4.4 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.4 | 3.0 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.4 | 1.8 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.4 | 4.6 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.4 | 2.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 5.0 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.3 | 1.0 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.3 | 1.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.3 | 6.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.3 | 3.6 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.3 | 4.7 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.3 | 2.9 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.3 | 1.3 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.3 | 4.5 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.3 | 1.0 | GO:0019230 | proprioception(GO:0019230) sensory system development(GO:0048880) |
| 0.2 | 2.0 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 2.3 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 1.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 11.9 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.2 | 2.6 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 0.8 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 0.9 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 0.9 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 1.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 0.9 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.2 | 3.4 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.2 | 4.4 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 1.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 1.4 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 2.8 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.1 | 3.6 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 1.4 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 | 5.2 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 3.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 1.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 3.4 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 2.7 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.1 | 9.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 7.4 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 0.9 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.5 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 2.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 2.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 1.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 1.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 4.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.4 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 2.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 3.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 1.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 1.5 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 0.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 13.4 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.1 | 0.2 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.1 | 2.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.9 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 3.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 5.3 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.7 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.5 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.3 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 1.1 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 1.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.6 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 3.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.3 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.2 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.9 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) circadian sleep/wake cycle, sleep(GO:0050802) |
| 0.0 | 1.4 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 6.7 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 1.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 2.4 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.1 | GO:0045333 | cellular respiration(GO:0045333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.5 | 46.6 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 8.9 | 133.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.7 | 11.9 | GO:0070695 | FHF complex(GO:0070695) |
| 1.3 | 10.4 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 1.1 | 26.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 1.1 | 9.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.0 | 14.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.8 | 2.4 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.6 | 53.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.5 | 1.6 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.5 | 2.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.5 | 7.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.5 | 1.8 | GO:0071942 | XPC complex(GO:0071942) |
| 0.4 | 25.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.3 | 3.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 45.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.3 | 3.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 5.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 15.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.2 | 1.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 6.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 47.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 9.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 20.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 16.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 7.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 8.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 10.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 36.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.9 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 2.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 1.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 6.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.8 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 2.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.9 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 8.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.8 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 2.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 12.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.8 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 2.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 15.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 4.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 11.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 10.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 7.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 10.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 2.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.9 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 3.0 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 45.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 8.4 | 25.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 5.1 | 133.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 5.0 | 40.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 3.8 | 26.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 3.8 | 22.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 3.2 | 9.6 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 3.2 | 25.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 2.9 | 17.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 2.7 | 8.1 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 2.7 | 8.0 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 2.6 | 12.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 2.5 | 17.4 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 2.4 | 9.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 2.1 | 15.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 2.0 | 8.2 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 1.9 | 7.8 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.9 | 7.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.8 | 5.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.7 | 61.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.6 | 6.3 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.5 | 4.5 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 1.5 | 42.2 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 1.4 | 17.9 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 1.2 | 10.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 1.0 | 3.1 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.9 | 4.5 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.9 | 2.7 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.9 | 4.4 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.9 | 4.3 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.9 | 6.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.7 | 15.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.7 | 2.1 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.7 | 11.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.7 | 11.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.7 | 2.6 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.6 | 1.9 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.6 | 13.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.6 | 3.5 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.5 | 14.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.5 | 3.6 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.5 | 4.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.5 | 4.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.4 | 1.3 | GO:0016232 | HNK-1 sulfotransferase activity(GO:0016232) |
| 0.4 | 2.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.4 | 3.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.4 | 3.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 2.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 1.0 | GO:0016715 | dopamine beta-monooxygenase activity(GO:0004500) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.3 | 1.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.3 | 34.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 3.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 4.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 0.9 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 1.8 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.2 | 0.9 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.2 | 2.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 9.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 34.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.2 | 2.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 3.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 7.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.2 | 20.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.2 | 1.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 2.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.9 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.9 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 2.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 4.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 10.4 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.1 | 1.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.1 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 0.1 | 0.5 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.3 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 4.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 23.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.7 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 1.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 2.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 2.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 2.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 4.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 6.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.9 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 1.1 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.5 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 1.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 2.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 10.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 9.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 125.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.3 | 20.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 16.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 4.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 2.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 11.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 26.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 8.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 4.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 6.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 5.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 7.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 2.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 2.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 36.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.3 | 181.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 1.3 | 17.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.3 | 45.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 1.3 | 59.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.8 | 13.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.7 | 8.7 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.5 | 10.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.5 | 6.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 6.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 19.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 13.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 4.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 4.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 4.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 7.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 17.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 7.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 3.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 4.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 15.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 6.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 3.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 9.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 8.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 6.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 0.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 5.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 3.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 3.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 2.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 7.4 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 1.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 3.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |