Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nr2e1
Z-value: 3.42
Transcription factors associated with Nr2e1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2e1
|
ENSMUSG00000019803.12 | nuclear receptor subfamily 2, group E, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2e1 | mm39_v1_chr10_-_42459624_42459635 | 0.00 | 9.8e-01 | Click! |
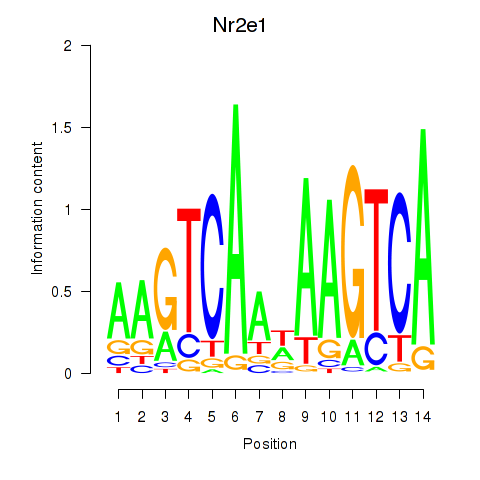
Activity profile of Nr2e1 motif
Sorted Z-values of Nr2e1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_62005498 | 109.86 |
ENSMUST00000107488.4
ENSMUST00000107472.8 ENSMUST00000084531.11 |
Mup3
|
major urinary protein 3 |
| chr4_-_60457902 | 107.44 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr4_-_60777462 | 91.90 |
ENSMUST00000211875.2
|
Mup22
|
major urinary protein 22 |
| chr4_-_61437704 | 87.64 |
ENSMUST00000095051.6
ENSMUST00000107483.8 |
Mup16
|
major urinary protein 16 |
| chr4_-_60697274 | 77.33 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr4_-_61259997 | 65.08 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chr4_-_61259801 | 59.68 |
ENSMUST00000125461.8
|
Mup14
|
major urinary protein 14 |
| chr4_-_62069046 | 48.36 |
ENSMUST00000077719.4
|
Mup21
|
major urinary protein 21 |
| chr6_-_141892517 | 39.25 |
ENSMUST00000168119.8
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr5_-_87288177 | 17.05 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr3_+_130411294 | 15.11 |
ENSMUST00000163620.8
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr13_+_24023428 | 13.98 |
ENSMUST00000091698.12
ENSMUST00000110422.3 ENSMUST00000166467.9 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr4_+_60003438 | 12.71 |
ENSMUST00000107517.8
ENSMUST00000107520.2 |
Mup6
|
major urinary protein 6 |
| chr13_+_24023386 | 10.81 |
ENSMUST00000039721.14
|
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr4_-_59960659 | 10.79 |
ENSMUST00000075973.3
|
Mup4
|
major urinary protein 4 |
| chr5_-_87054796 | 10.11 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr17_+_32904629 | 10.03 |
ENSMUST00000008801.7
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr17_+_32904601 | 9.77 |
ENSMUST00000168171.8
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr14_+_28740162 | 9.11 |
ENSMUST00000055662.4
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr8_+_36956345 | 8.84 |
ENSMUST00000171777.2
|
Trmt9b
|
tRNA methyltransferase 9B |
| chr11_+_68858942 | 8.26 |
ENSMUST00000102606.10
ENSMUST00000018884.6 |
Slc25a35
|
solute carrier family 25, member 35 |
| chr18_-_60881405 | 7.59 |
ENSMUST00000237070.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr3_+_96432479 | 7.01 |
ENSMUST00000049208.11
|
Hjv
|
hemojuvelin BMP co-receptor |
| chr18_-_60881679 | 6.23 |
ENSMUST00000237783.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr7_-_80055168 | 6.08 |
ENSMUST00000107362.10
ENSMUST00000135306.3 |
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr12_+_111132847 | 5.61 |
ENSMUST00000021706.11
|
Traf3
|
TNF receptor-associated factor 3 |
| chr12_+_111132779 | 5.09 |
ENSMUST00000117269.8
|
Traf3
|
TNF receptor-associated factor 3 |
| chr14_+_31750946 | 4.83 |
ENSMUST00000022460.11
|
Galnt15
|
polypeptide N-acetylgalactosaminyltransferase 15 |
| chr7_+_114367971 | 4.40 |
ENSMUST00000117543.3
ENSMUST00000151464.2 |
Insc
|
INSC spindle orientation adaptor protein |
| chr9_-_71070506 | 4.11 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr17_+_48037758 | 3.99 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr15_+_41694317 | 3.39 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr12_+_111132908 | 3.06 |
ENSMUST00000139162.8
ENSMUST00000060274.7 |
Traf3
|
TNF receptor-associated factor 3 |
| chr4_-_119272690 | 2.44 |
ENSMUST00000238287.2
ENSMUST00000238759.2 ENSMUST00000063642.10 |
Ccdc30
|
coiled-coil domain containing 30 |
| chrM_+_7006 | 2.31 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr7_+_131144596 | 2.26 |
ENSMUST00000046093.6
|
Hmx3
|
H6 homeobox 3 |
| chr12_+_86781141 | 2.14 |
ENSMUST00000223308.2
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr6_+_65567373 | 2.10 |
ENSMUST00000114236.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr2_-_22930149 | 2.08 |
ENSMUST00000091394.13
ENSMUST00000093171.13 |
Abi1
|
abl interactor 1 |
| chr6_+_42377172 | 2.08 |
ENSMUST00000057398.4
|
Tas2r143
|
taste receptor, type 2, member 143 |
| chr12_+_86781154 | 1.97 |
ENSMUST00000095527.6
|
Lrrc74a
|
leucine rich repeat containing 74A |
| chr8_+_71156071 | 1.93 |
ENSMUST00000212436.2
|
Iqcn
|
IQ motif containing N |
| chr3_+_14643669 | 1.84 |
ENSMUST00000029069.13
ENSMUST00000165922.3 |
E2f5
|
E2F transcription factor 5 |
| chr8_-_62355690 | 1.83 |
ENSMUST00000121785.9
ENSMUST00000034057.14 |
Palld
|
palladin, cytoskeletal associated protein |
| chr14_+_26414422 | 1.82 |
ENSMUST00000022433.12
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr4_-_119272640 | 1.78 |
ENSMUST00000238293.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr4_-_119272667 | 1.64 |
ENSMUST00000238609.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr4_+_33924632 | 1.62 |
ENSMUST00000057188.7
|
Cnr1
|
cannabinoid receptor 1 (brain) |
| chrX_-_105528503 | 1.31 |
ENSMUST00000138724.8
ENSMUST00000149331.2 |
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr6_+_120070307 | 1.31 |
ENSMUST00000112711.9
|
Ninj2
|
ninjurin 2 |
| chrY_+_24411927 | 1.28 |
ENSMUST00000179663.2
|
Gm20809
|
predicted gene, 20809 |
| chrY_+_68550741 | 1.28 |
ENSMUST00000177765.2
|
Gm20816
|
predicted gene, 20816 |
| chrY_-_65214536 | 1.28 |
ENSMUST00000177663.2
|
Gm20924
|
predicted gene, 20924 |
| chrY_-_77972959 | 1.25 |
ENSMUST00000178900.2
|
Gm20867
|
predicted gene, 20867 |
| chr9_-_42368880 | 1.23 |
ENSMUST00000125995.8
|
Tbcel
|
tubulin folding cofactor E-like |
| chr2_-_22930188 | 1.19 |
ENSMUST00000114544.10
ENSMUST00000139038.8 ENSMUST00000126112.8 ENSMUST00000178908.2 ENSMUST00000078977.14 ENSMUST00000140164.8 ENSMUST00000149719.8 |
Abi1
|
abl interactor 1 |
| chr2_-_22930104 | 1.17 |
ENSMUST00000153931.8
ENSMUST00000123948.8 |
Abi1
|
abl interactor 1 |
| chr3_-_146487102 | 1.08 |
ENSMUST00000005164.12
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chrY_-_9132561 | 1.07 |
ENSMUST00000171947.3
|
Gm21292
|
predicted gene, 21292 |
| chrY_+_21166083 | 1.00 |
ENSMUST00000178234.2
|
Gm20909
|
predicted gene, 20909 |
| chrY_-_53413354 | 1.00 |
ENSMUST00000179137.2
|
Gm20747
|
predicted gene, 20747 |
| chr5_-_123185073 | 0.94 |
ENSMUST00000031437.14
|
Morn3
|
MORN repeat containing 3 |
| chr7_+_104713682 | 0.94 |
ENSMUST00000213622.2
|
Olfr678
|
olfactory receptor 678 |
| chrY_-_79161056 | 0.92 |
ENSMUST00000179922.2
|
Gm20917
|
predicted gene, 20917 |
| chr9_+_8544143 | 0.81 |
ENSMUST00000050433.8
ENSMUST00000217462.2 |
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr3_-_24837772 | 0.78 |
ENSMUST00000203414.2
|
Naaladl2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr2_-_111820618 | 0.69 |
ENSMUST00000216948.2
ENSMUST00000214935.2 ENSMUST00000217452.2 ENSMUST00000215045.2 |
Olfr1309
|
olfactory receptor 1309 |
| chr10_+_18345706 | 0.67 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chrY_+_21242966 | 0.56 |
ENSMUST00000179095.2
|
Gm20865
|
predicted gene, 20865 |
| chr5_-_123185029 | 0.56 |
ENSMUST00000045843.15
|
Morn3
|
MORN repeat containing 3 |
| chrX_-_133012457 | 0.54 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr11_-_58521327 | 0.40 |
ENSMUST00000214132.2
|
Olfr323
|
olfactory receptor 323 |
| chr19_-_11261177 | 0.28 |
ENSMUST00000186937.7
ENSMUST00000067673.13 |
Ms4a5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr4_+_28813152 | 0.16 |
ENSMUST00000108194.9
ENSMUST00000108191.2 |
Epha7
|
Eph receptor A7 |
| chr2_-_120916316 | 0.15 |
ENSMUST00000028721.8
|
Tgm5
|
transglutaminase 5 |
| chr2_+_14828903 | 0.10 |
ENSMUST00000193800.6
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
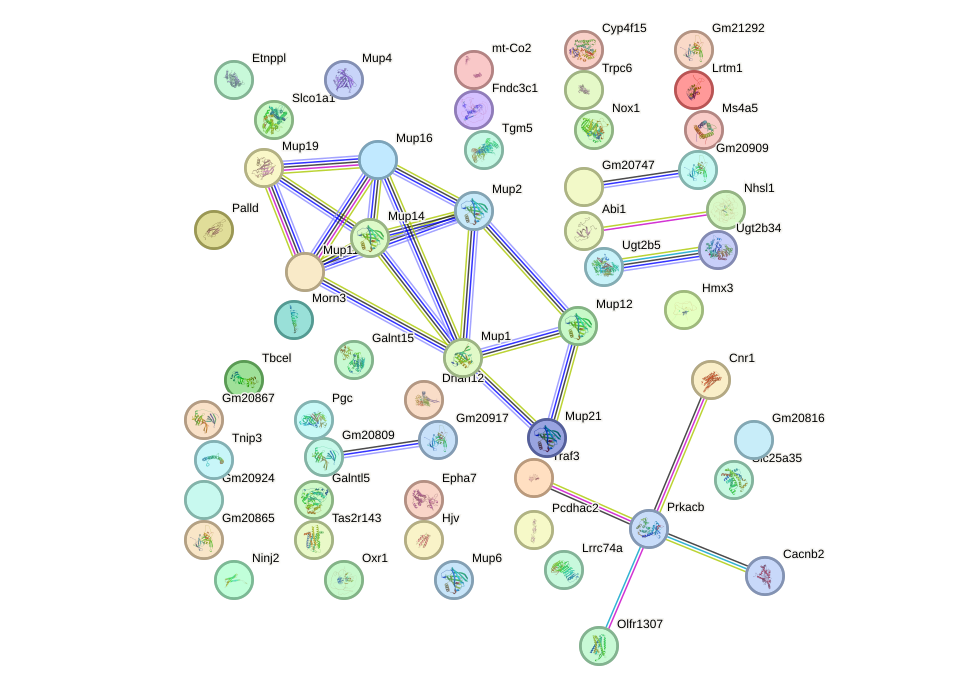
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2e1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.0 | 107.4 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 4.8 | 149.1 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 1.7 | 24.8 | GO:0015747 | urate transport(GO:0015747) |
| 1.5 | 13.8 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 1.5 | 6.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 1.3 | 4.0 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.3 | 13.8 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 1.0 | 4.1 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.5 | 1.6 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.2 | 3.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.5 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 4.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.1 | 2.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 2.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.8 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 9.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 4.4 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 1.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 1.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.7 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 2.3 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 2.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 8.3 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 1.3 | GO:0042246 | tissue regeneration(GO:0042246) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 13.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.3 | 6.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.3 | 4.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.8 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 2.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 226.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 43.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 24.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 25.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 4.4 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.8 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 1.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 4.8 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 18.5 | GO:0005739 | mitochondrion(GO:0005739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 35.8 | 107.4 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 5.0 | 15.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 2.5 | 24.8 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 2.0 | 13.8 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 1.4 | 118.1 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.9 | 13.8 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.8 | 35.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.8 | 4.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.7 | 2.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.7 | 27.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 1.6 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.3 | 6.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 4.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 4.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 19.8 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 4.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.5 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 2.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 9.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 1.8 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 2.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 3.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 5.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 6.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 13.8 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.4 | 6.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 13.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 4.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |