Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
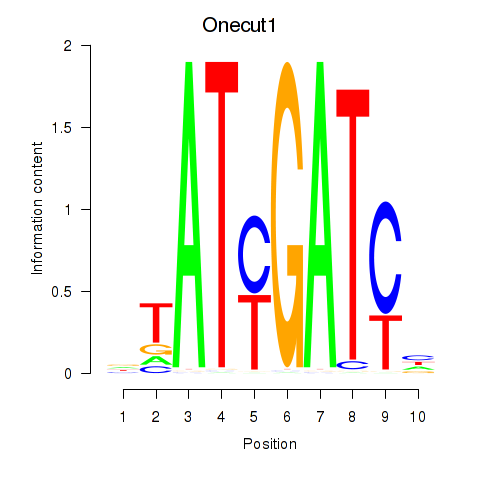
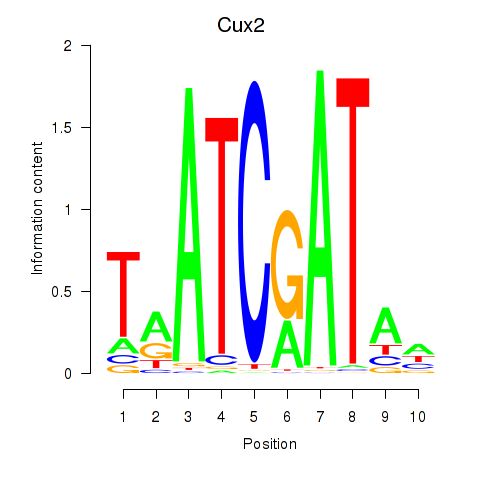
Results for Onecut1_Cux2
Z-value: 1.04
Transcription factors associated with Onecut1_Cux2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Onecut1
|
ENSMUSG00000043013.11 | one cut domain, family member 1 |
|
Cux2
|
ENSMUSG00000042589.19 | cut-like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Onecut1 | mm39_v1_chr9_+_74769166_74769203 | 0.55 | 4.7e-04 | Click! |
| Cux2 | mm39_v1_chr5_-_122187884_122187947 | -0.14 | 4.2e-01 | Click! |
Activity profile of Onecut1_Cux2 motif
Sorted Z-values of Onecut1_Cux2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_22769822 | 21.01 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr16_+_22769844 | 20.97 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr4_-_104733580 | 11.67 |
ENSMUST00000064873.9
ENSMUST00000106808.10 ENSMUST00000048947.15 |
C8a
|
complement component 8, alpha polypeptide |
| chr6_+_121323577 | 11.31 |
ENSMUST00000032200.16
|
Slc6a12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 |
| chr2_-_84605764 | 7.53 |
ENSMUST00000111641.2
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr2_-_84605732 | 7.53 |
ENSMUST00000023994.10
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| chr4_-_63072367 | 7.44 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr15_-_60793115 | 7.42 |
ENSMUST00000096418.5
|
A1bg
|
alpha-1-B glycoprotein |
| chr7_+_43856724 | 6.26 |
ENSMUST00000077354.5
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chr11_+_101258368 | 6.16 |
ENSMUST00000019469.3
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr1_+_172525613 | 5.96 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr3_+_146302832 | 5.90 |
ENSMUST00000029837.14
ENSMUST00000147409.2 ENSMUST00000121133.2 |
Uox
|
urate oxidase |
| chr11_+_78389913 | 5.68 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr3_+_28752050 | 5.10 |
ENSMUST00000029240.14
|
Slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr14_-_30665232 | 4.74 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr3_+_94280101 | 4.38 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr11_-_59927688 | 4.14 |
ENSMUST00000102692.10
|
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr2_-_147888816 | 3.92 |
ENSMUST00000172928.2
ENSMUST00000047315.10 |
Foxa2
|
forkhead box A2 |
| chr15_-_78352801 | 3.43 |
ENSMUST00000229124.2
ENSMUST00000230226.2 ENSMUST00000017086.5 |
Tmprss6
|
transmembrane serine protease 6 |
| chr4_-_46991842 | 3.42 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr15_+_54975713 | 3.31 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr8_+_127790772 | 3.17 |
ENSMUST00000079777.12
ENSMUST00000160272.8 ENSMUST00000162907.8 ENSMUST00000162536.8 ENSMUST00000026921.13 ENSMUST00000162665.8 ENSMUST00000162602.8 ENSMUST00000160581.8 ENSMUST00000161355.8 ENSMUST00000162531.8 ENSMUST00000160766.8 ENSMUST00000159537.8 |
Pard3
|
par-3 family cell polarity regulator |
| chr6_-_23132977 | 3.09 |
ENSMUST00000031707.14
|
Aass
|
aminoadipate-semialdehyde synthase |
| chr8_+_56747613 | 2.94 |
ENSMUST00000034026.10
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD) |
| chr7_+_67297152 | 2.64 |
ENSMUST00000032774.16
ENSMUST00000107471.8 |
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr15_-_96929086 | 2.58 |
ENSMUST00000230086.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr7_+_67305162 | 2.43 |
ENSMUST00000107470.2
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr6_-_136899167 | 2.38 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr5_+_130413532 | 2.33 |
ENSMUST00000202728.2
|
Caln1
|
calneuron 1 |
| chr3_-_146302343 | 2.20 |
ENSMUST00000029836.9
|
Dnase2b
|
deoxyribonuclease II beta |
| chr1_+_191449946 | 2.20 |
ENSMUST00000133076.7
ENSMUST00000110855.8 |
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chrX_+_100420873 | 2.06 |
ENSMUST00000052130.14
|
Gjb1
|
gap junction protein, beta 1 |
| chr19_-_38113696 | 2.05 |
ENSMUST00000025951.14
ENSMUST00000237287.2 |
Rbp4
|
retinol binding protein 4, plasma |
| chr5_+_3646066 | 1.89 |
ENSMUST00000006061.13
ENSMUST00000121291.8 ENSMUST00000142516.2 |
Pex1
|
peroxisomal biogenesis factor 1 |
| chr15_+_54975814 | 1.87 |
ENSMUST00000100660.11
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr14_-_55329734 | 1.65 |
ENSMUST00000036328.9
|
Zfhx2
|
zinc finger homeobox 2 |
| chr19_+_41017714 | 1.48 |
ENSMUST00000051806.12
ENSMUST00000112200.3 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chrM_+_14138 | 1.46 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr12_+_55646209 | 1.39 |
ENSMUST00000051857.5
|
Insm2
|
insulinoma-associated 2 |
| chr9_-_114673158 | 1.39 |
ENSMUST00000047013.4
|
Cmtm8
|
CKLF-like MARVEL transmembrane domain containing 8 |
| chr19_+_56276343 | 1.38 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr18_+_36098090 | 1.29 |
ENSMUST00000176873.8
ENSMUST00000177432.8 ENSMUST00000175734.2 |
Psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr19_-_57185988 | 1.27 |
ENSMUST00000099294.9
|
Ablim1
|
actin-binding LIM protein 1 |
| chr19_-_57185928 | 1.16 |
ENSMUST00000111544.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr14_-_4488167 | 1.11 |
ENSMUST00000022304.12
|
Thrb
|
thyroid hormone receptor beta |
| chr19_+_31060237 | 1.11 |
ENSMUST00000066039.8
|
Cstf2t
|
cleavage stimulation factor, 3' pre-RNA subunit 2, tau |
| chr10_+_29019645 | 1.11 |
ENSMUST00000092629.4
|
Soga3
|
SOGA family member 3 |
| chr12_-_25146078 | 1.07 |
ENSMUST00000222667.2
ENSMUST00000020974.7 |
Id2
|
inhibitor of DNA binding 2 |
| chr19_-_57185808 | 1.06 |
ENSMUST00000111546.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chrM_-_14061 | 0.98 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr5_+_3646355 | 0.98 |
ENSMUST00000195894.2
|
Pex1
|
peroxisomal biogenesis factor 1 |
| chr8_-_13250535 | 0.91 |
ENSMUST00000165605.4
ENSMUST00000209691.2 ENSMUST00000211128.2 ENSMUST00000210317.2 |
Grtp1
|
GH regulated TBC protein 1 |
| chr19_-_37155410 | 0.71 |
ENSMUST00000133988.2
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr1_-_13197387 | 0.66 |
ENSMUST00000047577.7
|
Prdm14
|
PR domain containing 14 |
| chr5_+_92831150 | 0.52 |
ENSMUST00000113055.9
|
Shroom3
|
shroom family member 3 |
| chr3_+_5283577 | 0.51 |
ENSMUST00000175866.8
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr3_-_79749949 | 0.50 |
ENSMUST00000029568.7
|
Tmem144
|
transmembrane protein 144 |
| chr13_-_54836059 | 0.47 |
ENSMUST00000122935.2
ENSMUST00000128257.8 |
Rnf44
|
ring finger protein 44 |
| chr3_+_5283606 | 0.47 |
ENSMUST00000026284.13
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr2_+_163444248 | 0.45 |
ENSMUST00000152135.8
|
Ttpal
|
tocopherol (alpha) transfer protein-like |
| chr19_+_26728339 | 0.42 |
ENSMUST00000175953.3
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_3646071 | 0.42 |
ENSMUST00000121877.3
|
Rbm48
|
RNA binding motif protein 48 |
| chr9_+_59496571 | 0.42 |
ENSMUST00000121266.8
ENSMUST00000118164.3 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chr19_-_57185861 | 0.40 |
ENSMUST00000111550.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr6_+_140378688 | 0.38 |
ENSMUST00000203774.3
|
Plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr10_+_76089674 | 0.36 |
ENSMUST00000036387.8
|
S100b
|
S100 protein, beta polypeptide, neural |
| chr16_-_95260104 | 0.34 |
ENSMUST00000176345.10
ENSMUST00000121809.11 ENSMUST00000233664.2 ENSMUST00000122199.10 |
Erg
|
ETS transcription factor |
| chr5_+_92831468 | 0.33 |
ENSMUST00000168878.8
|
Shroom3
|
shroom family member 3 |
| chr15_-_8739893 | 0.28 |
ENSMUST00000157065.2
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr11_-_120515799 | 0.25 |
ENSMUST00000106183.3
ENSMUST00000080202.12 |
Sirt7
|
sirtuin 7 |
| chr4_-_97472844 | 0.24 |
ENSMUST00000107067.8
ENSMUST00000107068.9 |
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr2_+_129642371 | 0.22 |
ENSMUST00000165413.9
ENSMUST00000166282.3 |
Stk35
|
serine/threonine kinase 35 |
| chr8_+_114369838 | 0.21 |
ENSMUST00000095173.3
ENSMUST00000034219.12 ENSMUST00000212269.2 |
Syce1l
|
synaptonemal complex central element protein 1 like |
| chr10_-_63926044 | 0.20 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr5_+_63806451 | 0.19 |
ENSMUST00000159584.3
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr15_+_92059224 | 0.19 |
ENSMUST00000068378.6
|
Cntn1
|
contactin 1 |
| chr19_-_32038838 | 0.18 |
ENSMUST00000096119.5
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr1_+_88334678 | 0.18 |
ENSMUST00000027518.12
|
Spp2
|
secreted phosphoprotein 2 |
| chr15_+_22549108 | 0.16 |
ENSMUST00000163361.8
|
Cdh18
|
cadherin 18 |
| chr18_-_72484126 | 0.15 |
ENSMUST00000114943.11
|
Dcc
|
deleted in colorectal carcinoma |
| chr2_-_60503998 | 0.15 |
ENSMUST00000059888.15
ENSMUST00000154764.2 |
Itgb6
|
integrin beta 6 |
| chr2_-_7086018 | 0.15 |
ENSMUST00000114923.3
ENSMUST00000182706.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chrX_-_50294867 | 0.14 |
ENSMUST00000114876.9
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr15_-_8740218 | 0.14 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr12_-_46865709 | 0.13 |
ENSMUST00000021438.8
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr2_+_147206910 | 0.11 |
ENSMUST00000109968.3
|
Pax1
|
paired box 1 |
| chr8_+_59364789 | 0.10 |
ENSMUST00000062978.7
|
BC030500
|
cDNA sequence BC030500 |
| chr12_+_72488625 | 0.10 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr7_-_105460172 | 0.09 |
ENSMUST00000033170.7
ENSMUST00000176498.2 ENSMUST00000176887.2 ENSMUST00000124482.3 |
Mrpl17
|
mitochondrial ribosomal protein L17 |
| chr3_+_68479578 | 0.08 |
ENSMUST00000170788.9
|
Schip1
|
schwannomin interacting protein 1 |
| chr8_-_32408380 | 0.08 |
ENSMUST00000208497.3
ENSMUST00000207584.3 |
Nrg1
|
neuregulin 1 |
| chr6_+_40468167 | 0.07 |
ENSMUST00000064932.6
|
Tas2r137
|
taste receptor, type 2, member 137 |
| chr8_+_23114035 | 0.04 |
ENSMUST00000033936.8
|
Dkk4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr5_-_3646540 | 0.03 |
ENSMUST00000042753.14
|
Rbm48
|
RNA binding motif protein 48 |
| chr1_+_59621555 | 0.03 |
ENSMUST00000212835.2
|
Gm973
|
predicted gene 973 |
| chr9_-_29874401 | 0.02 |
ENSMUST00000075069.11
|
Ntm
|
neurotrimin |
| chr3_-_33136153 | 0.01 |
ENSMUST00000108225.10
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
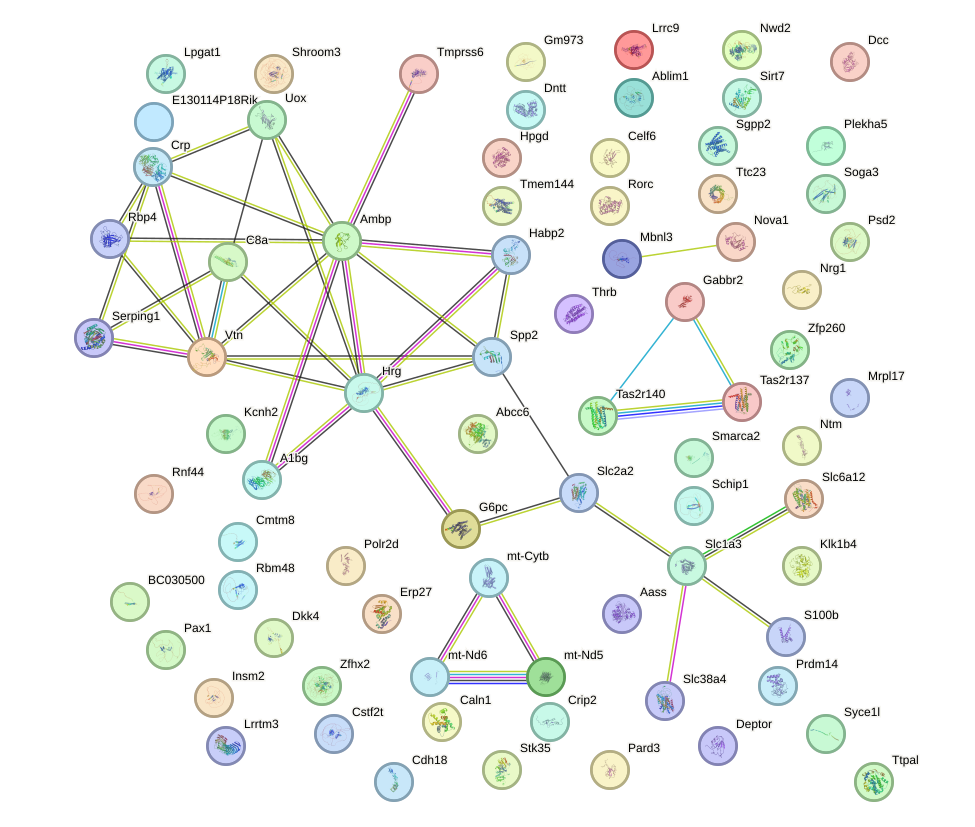
Network of associatons between targets according to the STRING database.
First level regulatory network of Onecut1_Cux2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.0 | 42.0 | GO:0097037 | heme export(GO:0097037) |
| 5.0 | 15.1 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 2.3 | 11.3 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 1.3 | 3.9 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 1.0 | 3.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 1.0 | 5.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 1.0 | 2.9 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.7 | 6.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.7 | 11.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.6 | 6.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.6 | 4.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.5 | 7.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.5 | 3.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.4 | 5.7 | GO:0097421 | smooth muscle cell-matrix adhesion(GO:0061302) liver regeneration(GO:0097421) |
| 0.4 | 2.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.4 | 4.4 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.4 | 1.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.3 | 5.9 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 0.7 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 | 1.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.2 | 5.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 2.9 | GO:0007567 | parturition(GO:0007567) |
| 0.2 | 1.5 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.2 | 2.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.2 | 3.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 6.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.2 | 0.7 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.1 | 1.4 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 4.7 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 2.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 3.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.4 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.1 | 0.3 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 2.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 1.5 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 1.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 3.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.5 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.2 | GO:0007128 | meiotic prophase I(GO:0007128) prophase(GO:0051324) |
| 0.0 | 0.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.0 | 42.0 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 1.3 | 11.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.8 | 5.7 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.7 | 3.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.4 | 3.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.5 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.1 | 33.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 2.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 9.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 2.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 6.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 8.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 6.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.5 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.4 | GO:0019862 | IgA binding(GO:0019862) |
| 1.6 | 11.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 1.5 | 6.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.2 | 32.7 | GO:0001848 | complement binding(GO:0001848) |
| 1.0 | 5.1 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.7 | 4.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.7 | 2.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.6 | 3.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.5 | 5.9 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.4 | 2.2 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.4 | 2.9 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.4 | 4.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.4 | 42.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 5.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 1.5 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 2.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 3.1 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.1 | GO:0070324 | thyroid hormone receptor activity(GO:0004887) thyroid hormone binding(GO:0070324) |
| 0.1 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 3.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 6.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 2.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 4.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 70.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 5.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 15.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 7.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 6.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.7 | 15.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.5 | 11.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 11.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 40.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 5.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 3.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 4.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 3.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 6.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 3.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 5.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 5.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 3.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |