Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
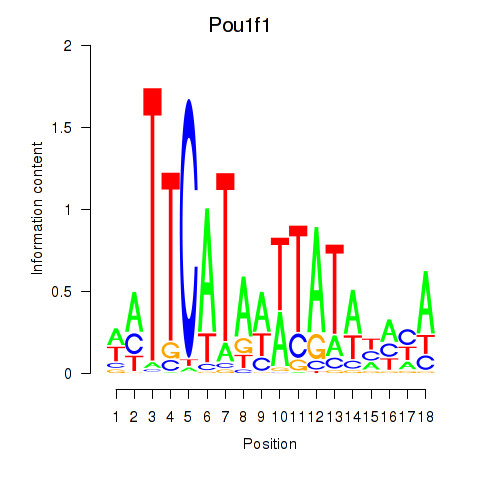
Results for Pou1f1
Z-value: 3.72
Transcription factors associated with Pou1f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou1f1
|
ENSMUSG00000004842.19 | POU domain, class 1, transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou1f1 | mm39_v1_chr16_+_65317389_65317434 | -0.15 | 3.7e-01 | Click! |
Activity profile of Pou1f1 motif
Sorted Z-values of Pou1f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_87074380 | 72.86 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr5_-_87485023 | 52.30 |
ENSMUST00000031195.3
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr4_-_60457902 | 52.16 |
ENSMUST00000084548.11
ENSMUST00000103012.10 ENSMUST00000107499.4 |
Mup1
|
major urinary protein 1 |
| chr4_-_60777462 | 51.95 |
ENSMUST00000211875.2
|
Mup22
|
major urinary protein 22 |
| chr4_-_60697274 | 48.09 |
ENSMUST00000117932.2
|
Mup12
|
major urinary protein 12 |
| chr4_-_61259997 | 47.31 |
ENSMUST00000071005.9
ENSMUST00000075206.12 |
Mup14
|
major urinary protein 14 |
| chr4_-_60222580 | 46.32 |
ENSMUST00000095058.5
ENSMUST00000163931.8 |
Mup8
|
major urinary protein 8 |
| chr8_-_94006345 | 36.95 |
ENSMUST00000034178.9
|
Ces1f
|
carboxylesterase 1F |
| chr4_-_61437704 | 36.61 |
ENSMUST00000095051.6
ENSMUST00000107483.8 |
Mup16
|
major urinary protein 16 |
| chr4_-_60538151 | 35.29 |
ENSMUST00000098047.3
|
Mup10
|
major urinary protein 10 |
| chr4_-_61592331 | 34.54 |
ENSMUST00000098040.4
|
Mup18
|
major urinary protein 18 |
| chr5_-_87240405 | 34.25 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr19_+_39275518 | 34.18 |
ENSMUST00000003137.15
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr4_-_60377932 | 31.34 |
ENSMUST00000107506.9
ENSMUST00000122381.8 ENSMUST00000118759.8 ENSMUST00000132829.3 |
Mup9
|
major urinary protein 9 |
| chr4_-_61972348 | 31.33 |
ENSMUST00000074018.4
|
Mup20
|
major urinary protein 20 |
| chr19_-_39637489 | 31.25 |
ENSMUST00000067328.7
|
Cyp2c67
|
cytochrome P450, family 2, subfamily c, polypeptide 67 |
| chr13_+_4486105 | 31.05 |
ENSMUST00000156277.2
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr6_+_121815473 | 30.90 |
ENSMUST00000032228.9
|
Mug1
|
murinoglobulin 1 |
| chr5_-_87288177 | 30.50 |
ENSMUST00000067790.7
|
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr1_-_139708906 | 27.59 |
ENSMUST00000111986.8
ENSMUST00000027612.11 ENSMUST00000111989.9 |
Cfhr4
|
complement factor H-related 4 |
| chr4_-_60139857 | 27.38 |
ENSMUST00000107490.5
ENSMUST00000074700.9 |
Mup2
|
major urinary protein 2 |
| chr4_-_61700450 | 27.09 |
ENSMUST00000107477.2
ENSMUST00000080606.9 |
Mup19
|
major urinary protein 19 |
| chr4_-_96552349 | 26.31 |
ENSMUST00000030299.8
|
Cyp2j5
|
cytochrome P450, family 2, subfamily j, polypeptide 5 |
| chr19_+_40078132 | 24.24 |
ENSMUST00000068094.13
ENSMUST00000080171.3 |
Cyp2c50
|
cytochrome P450, family 2, subfamily c, polypeptide 50 |
| chr5_-_87572060 | 23.71 |
ENSMUST00000072818.6
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr4_-_60618357 | 23.31 |
ENSMUST00000084544.5
ENSMUST00000098046.10 |
Mup11
|
major urinary protein 11 |
| chr3_+_138121245 | 21.77 |
ENSMUST00000161312.8
ENSMUST00000013458.9 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr8_-_93956143 | 21.52 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr8_+_110717062 | 21.43 |
ENSMUST00000001720.14
ENSMUST00000143741.2 |
Tat
|
tyrosine aminotransferase |
| chr4_-_61357980 | 21.21 |
ENSMUST00000095049.5
|
Mup15
|
major urinary protein 15 |
| chr5_-_145816774 | 20.76 |
ENSMUST00000035918.8
|
Cyp3a11
|
cytochrome P450, family 3, subfamily a, polypeptide 11 |
| chr19_-_40062174 | 20.60 |
ENSMUST00000048959.5
|
Cyp2c54
|
cytochrome P450, family 2, subfamily c, polypeptide 54 |
| chr6_-_55152002 | 20.51 |
ENSMUST00000003569.6
|
Inmt
|
indolethylamine N-methyltransferase |
| chr3_-_98537568 | 20.02 |
ENSMUST00000044094.6
|
Hsd3b5
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 |
| chr6_+_121983720 | 19.02 |
ENSMUST00000081777.8
|
Mug2
|
murinoglobulin 2 |
| chr19_+_39980868 | 18.84 |
ENSMUST00000049178.3
|
Cyp2c37
|
cytochrome P450, family 2. subfamily c, polypeptide 37 |
| chr15_+_4756684 | 17.70 |
ENSMUST00000161997.8
ENSMUST00000022788.15 |
C6
|
complement component 6 |
| chr19_-_39451509 | 17.10 |
ENSMUST00000035488.3
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr15_+_4756657 | 16.61 |
ENSMUST00000162585.8
|
C6
|
complement component 6 |
| chr10_-_127843377 | 15.54 |
ENSMUST00000219447.2
ENSMUST00000219780.2 ENSMUST00000219707.2 ENSMUST00000219953.2 ENSMUST00000219183.2 |
Hsd17b6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr15_+_6474808 | 15.30 |
ENSMUST00000022749.17
ENSMUST00000239466.2 |
C9
|
complement component 9 |
| chr3_+_59989282 | 15.18 |
ENSMUST00000029326.6
|
Sucnr1
|
succinate receptor 1 |
| chr14_+_66207163 | 15.09 |
ENSMUST00000153460.8
|
Clu
|
clusterin |
| chr13_+_4484305 | 15.01 |
ENSMUST00000021630.15
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr7_+_140343652 | 14.63 |
ENSMUST00000026552.9
ENSMUST00000209253.2 ENSMUST00000210235.2 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr1_+_58152295 | 14.55 |
ENSMUST00000040999.14
ENSMUST00000162011.3 |
Aox3
|
aldehyde oxidase 3 |
| chr3_+_94600863 | 14.02 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr19_-_40175709 | 13.68 |
ENSMUST00000051846.13
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr5_-_89583469 | 13.37 |
ENSMUST00000200534.2
|
Gc
|
vitamin D binding protein |
| chr8_-_45786226 | 12.79 |
ENSMUST00000095328.6
|
Cyp4v3
|
cytochrome P450, family 4, subfamily v, polypeptide 3 |
| chr1_+_21310803 | 12.65 |
ENSMUST00000027067.15
|
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr5_+_115061293 | 12.40 |
ENSMUST00000031540.11
ENSMUST00000112143.4 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr19_-_4092218 | 12.11 |
ENSMUST00000237999.2
ENSMUST00000042700.12 |
Gstp2
|
glutathione S-transferase, pi 2 |
| chr10_+_87357657 | 11.98 |
ENSMUST00000020241.17
|
Pah
|
phenylalanine hydroxylase |
| chr1_+_21310821 | 11.90 |
ENSMUST00000121676.8
ENSMUST00000124990.3 |
Gsta3
|
glutathione S-transferase, alpha 3 |
| chr7_+_25597045 | 11.77 |
ENSMUST00000072438.13
ENSMUST00000005477.6 |
Cyp2b10
|
cytochrome P450, family 2, subfamily b, polypeptide 10 |
| chr11_-_11848107 | 11.15 |
ENSMUST00000178704.8
|
Ddc
|
dopa decarboxylase |
| chr5_+_45650716 | 10.99 |
ENSMUST00000046122.11
|
Lap3
|
leucine aminopeptidase 3 |
| chr10_-_126956991 | 10.05 |
ENSMUST00000080975.6
ENSMUST00000164259.9 |
Os9
|
amplified in osteosarcoma |
| chr1_+_58069090 | 9.92 |
ENSMUST00000001027.7
|
Aox1
|
aldehyde oxidase 1 |
| chr8_-_94063823 | 9.64 |
ENSMUST00000044602.8
|
Ces1g
|
carboxylesterase 1G |
| chr6_-_141892517 | 9.16 |
ENSMUST00000168119.8
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr2_-_110136074 | 9.12 |
ENSMUST00000046233.9
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr7_-_46382003 | 9.05 |
ENSMUST00000006952.9
|
Saa4
|
serum amyloid A 4 |
| chr5_+_45650821 | 8.75 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr12_-_103423472 | 8.74 |
ENSMUST00000044687.7
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr10_-_115198093 | 8.68 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chr3_+_62245765 | 8.54 |
ENSMUST00000079300.13
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr9_-_121745354 | 8.26 |
ENSMUST00000062474.5
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chrX_-_37545311 | 8.23 |
ENSMUST00000074913.12
ENSMUST00000016678.14 ENSMUST00000061755.9 |
Lamp2
|
lysosomal-associated membrane protein 2 |
| chr9_+_78164402 | 8.00 |
ENSMUST00000217203.2
|
Gm3776
|
predicted gene 3776 |
| chr13_+_4283729 | 7.99 |
ENSMUST00000081326.7
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chrX_-_74886791 | 7.93 |
ENSMUST00000137192.2
|
Pls3
|
plastin 3 (T-isoform) |
| chrX_-_74886623 | 7.71 |
ENSMUST00000114057.8
|
Pls3
|
plastin 3 (T-isoform) |
| chr1_-_121260274 | 7.45 |
ENSMUST00000161068.2
|
Insig2
|
insulin induced gene 2 |
| chr1_+_185064339 | 7.36 |
ENSMUST00000027916.13
ENSMUST00000210277.2 ENSMUST00000151769.2 ENSMUST00000110965.2 |
Bpnt1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr6_+_138117519 | 7.34 |
ENSMUST00000120939.8
ENSMUST00000204628.3 ENSMUST00000140932.2 ENSMUST00000120302.8 |
Mgst1
|
microsomal glutathione S-transferase 1 |
| chrX_+_10118544 | 7.04 |
ENSMUST00000049910.13
|
Otc
|
ornithine transcarbamylase |
| chr7_-_100306160 | 7.00 |
ENSMUST00000107046.8
ENSMUST00000107045.9 ENSMUST00000139708.9 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr1_-_72323464 | 6.99 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr1_+_88128323 | 6.97 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr1_+_180878797 | 6.95 |
ENSMUST00000036819.7
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr1_-_72323407 | 6.92 |
ENSMUST00000097698.5
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr10_-_75616788 | 6.75 |
ENSMUST00000173512.2
ENSMUST00000173537.2 |
Gm20441
Gstt3
|
predicted gene 20441 glutathione S-transferase, theta 3 |
| chr19_+_39499288 | 6.75 |
ENSMUST00000025968.5
|
Cyp2c39
|
cytochrome P450, family 2, subfamily c, polypeptide 39 |
| chr15_-_77283286 | 6.66 |
ENSMUST00000175919.8
ENSMUST00000176074.9 |
Apol7a
|
apolipoprotein L 7a |
| chr4_-_118667141 | 6.54 |
ENSMUST00000084313.5
ENSMUST00000219094.2 |
Olfr1335
|
olfactory receptor 1335 |
| chr5_-_87402659 | 6.44 |
ENSMUST00000075858.4
|
Ugt2b37
|
UDP glucuronosyltransferase 2 family, polypeptide B37 |
| chr1_+_88093726 | 6.38 |
ENSMUST00000097659.5
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr11_-_11848044 | 6.37 |
ENSMUST00000066237.10
|
Ddc
|
dopa decarboxylase |
| chrX_+_10118600 | 6.20 |
ENSMUST00000115528.3
|
Otc
|
ornithine transcarbamylase |
| chr19_+_30210320 | 6.19 |
ENSMUST00000025797.7
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chr10_-_24712034 | 6.17 |
ENSMUST00000218044.2
ENSMUST00000020169.9 |
Enpp3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chrM_+_9459 | 6.13 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr4_+_60003438 | 6.10 |
ENSMUST00000107517.8
ENSMUST00000107520.2 |
Mup6
|
major urinary protein 6 |
| chr1_+_87998487 | 6.09 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr17_-_32643067 | 6.06 |
ENSMUST00000237130.2
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr5_-_139799953 | 6.06 |
ENSMUST00000044002.10
|
Tmem184a
|
transmembrane protein 184a |
| chr4_+_115375461 | 6.03 |
ENSMUST00000058785.10
ENSMUST00000094886.4 |
Cyp4a10
|
cytochrome P450, family 4, subfamily a, polypeptide 10 |
| chr6_-_138013901 | 6.00 |
ENSMUST00000150278.3
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr3_-_98660781 | 5.93 |
ENSMUST00000094050.11
ENSMUST00000090743.13 |
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 |
| chr4_+_134124691 | 5.90 |
ENSMUST00000105870.8
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chrM_+_8603 | 5.74 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr6_-_130314465 | 5.66 |
ENSMUST00000088017.5
ENSMUST00000111998.9 |
Klra3
|
killer cell lectin-like receptor, subfamily A, member 3 |
| chr10_-_75617245 | 5.57 |
ENSMUST00000001715.10
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr5_-_137919873 | 5.55 |
ENSMUST00000031741.8
|
Cyp3a13
|
cytochrome P450, family 3, subfamily a, polypeptide 13 |
| chr10_+_53213763 | 5.53 |
ENSMUST00000219491.2
ENSMUST00000163319.9 ENSMUST00000220197.2 ENSMUST00000046221.8 ENSMUST00000218468.2 ENSMUST00000219921.2 |
Pln
|
phospholamban |
| chr15_-_98575332 | 5.44 |
ENSMUST00000120997.2
ENSMUST00000109149.9 ENSMUST00000003451.11 |
Rnd1
|
Rho family GTPase 1 |
| chr3_+_94280101 | 5.39 |
ENSMUST00000029795.10
|
Rorc
|
RAR-related orphan receptor gamma |
| chr2_-_62242562 | 5.38 |
ENSMUST00000047812.8
|
Dpp4
|
dipeptidylpeptidase 4 |
| chr3_-_113367891 | 5.38 |
ENSMUST00000142505.9
|
Amy1
|
amylase 1, salivary |
| chr19_-_7780025 | 5.37 |
ENSMUST00000065634.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr1_-_140111018 | 5.30 |
ENSMUST00000192880.6
ENSMUST00000111977.8 |
Cfh
|
complement component factor h |
| chr6_+_40619913 | 5.19 |
ENSMUST00000238599.2
|
Mgam
|
maltase-glucoamylase |
| chrM_+_3906 | 5.17 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr10_+_87357816 | 5.09 |
ENSMUST00000218573.2
|
Pah
|
phenylalanine hydroxylase |
| chr10_+_87357782 | 4.96 |
ENSMUST00000219813.2
|
Pah
|
phenylalanine hydroxylase |
| chr18_-_75094323 | 4.95 |
ENSMUST00000066532.5
|
Lipg
|
lipase, endothelial |
| chr8_+_56747613 | 4.95 |
ENSMUST00000034026.10
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD) |
| chr13_-_47196607 | 4.91 |
ENSMUST00000124948.2
|
Tpmt
|
thiopurine methyltransferase |
| chr1_-_184543367 | 4.84 |
ENSMUST00000048462.13
ENSMUST00000110992.9 |
Mtarc1
|
mitochondrial amidoxime reducing component 1 |
| chrX_+_20416019 | 4.82 |
ENSMUST00000023832.7
|
Rgn
|
regucalcin |
| chr5_-_130053120 | 4.77 |
ENSMUST00000161640.8
ENSMUST00000161884.2 ENSMUST00000161094.8 |
Asl
|
argininosuccinate lyase |
| chr18_+_50261268 | 4.71 |
ENSMUST00000025385.7
|
Hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr6_-_3988835 | 4.70 |
ENSMUST00000203257.2
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr17_+_46807637 | 4.63 |
ENSMUST00000046497.8
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr8_+_36956345 | 4.57 |
ENSMUST00000171777.2
|
Trmt9b
|
tRNA methyltransferase 9B |
| chr3_+_93462387 | 4.56 |
ENSMUST00000045756.14
|
S100a10
|
S100 calcium binding protein A10 (calpactin) |
| chr15_-_100579813 | 4.54 |
ENSMUST00000230572.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr6_-_72593983 | 4.54 |
ENSMUST00000070524.5
|
Tgoln1
|
trans-golgi network protein |
| chr19_-_7779943 | 4.48 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr15_-_76193955 | 4.44 |
ENSMUST00000210024.2
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr11_+_101258368 | 4.36 |
ENSMUST00000019469.3
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr6_-_119365632 | 4.35 |
ENSMUST00000169744.8
|
Adipor2
|
adiponectin receptor 2 |
| chr6_+_71176811 | 4.32 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr5_-_147259245 | 4.28 |
ENSMUST00000100433.5
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
| chr13_+_4278681 | 4.24 |
ENSMUST00000118663.9
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr3_+_81906768 | 4.23 |
ENSMUST00000107736.2
|
Asic5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chr19_-_4489415 | 4.18 |
ENSMUST00000235680.2
ENSMUST00000117462.2 ENSMUST00000048197.10 |
Rhod
|
ras homolog family member D |
| chrM_+_9870 | 4.16 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr1_+_88022776 | 4.16 |
ENSMUST00000150634.8
ENSMUST00000058237.14 |
Ugt1a7c
|
UDP glucuronosyltransferase 1 family, polypeptide A7C |
| chr3_+_129630380 | 4.13 |
ENSMUST00000077918.7
|
Cfi
|
complement component factor i |
| chr11_+_66915969 | 4.10 |
ENSMUST00000079077.12
ENSMUST00000061786.6 |
Tmem220
|
transmembrane protein 220 |
| chr12_+_87204374 | 4.09 |
ENSMUST00000222222.2
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr1_-_140111138 | 4.09 |
ENSMUST00000111976.9
ENSMUST00000066859.13 |
Cfh
|
complement component factor h |
| chr18_+_84869456 | 4.04 |
ENSMUST00000160180.9
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr5_-_147831610 | 4.02 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr8_+_27532623 | 4.00 |
ENSMUST00000209856.2
ENSMUST00000098851.12 ENSMUST00000211393.2 ENSMUST00000211518.2 |
Plpbp
|
pyridoxal phosphate binding protein |
| chr12_-_103925197 | 3.97 |
ENSMUST00000122229.8
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr8_+_47246534 | 3.96 |
ENSMUST00000210218.2
|
Irf2
|
interferon regulatory factor 2 |
| chr1_+_87983099 | 3.96 |
ENSMUST00000138182.8
ENSMUST00000113142.10 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr16_-_35589726 | 3.95 |
ENSMUST00000023554.9
|
Slc49a4
|
solute carrier family 49 member 4 |
| chr10_-_115197775 | 3.94 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr5_-_87054796 | 3.88 |
ENSMUST00000031181.16
ENSMUST00000113333.2 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr9_+_78099229 | 3.86 |
ENSMUST00000034903.7
|
Gsta4
|
glutathione S-transferase, alpha 4 |
| chr13_+_113171645 | 3.85 |
ENSMUST00000180543.8
ENSMUST00000181568.8 ENSMUST00000109244.9 ENSMUST00000181117.8 ENSMUST00000181741.2 |
Cdc20b
|
cell division cycle 20B |
| chr10_+_75242745 | 3.80 |
ENSMUST00000039925.8
|
Upb1
|
ureidopropionase, beta |
| chr7_-_37927399 | 3.77 |
ENSMUST00000098513.6
|
Plekhf1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr11_+_96920751 | 3.75 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr6_-_128503666 | 3.72 |
ENSMUST00000143664.2
ENSMUST00000112132.8 |
Pzp
|
PZP, alpha-2-macroglobulin like |
| chr6_+_149043011 | 3.72 |
ENSMUST00000179873.8
ENSMUST00000047531.16 ENSMUST00000111548.8 ENSMUST00000111547.2 ENSMUST00000134306.8 ENSMUST00000147934.4 |
Etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr1_-_72251466 | 3.70 |
ENSMUST00000048860.9
|
Mreg
|
melanoregulin |
| chr7_+_100966289 | 3.65 |
ENSMUST00000163799.9
ENSMUST00000164479.9 |
Stard10
|
START domain containing 10 |
| chr3_+_94840352 | 3.61 |
ENSMUST00000090839.12
|
Selenbp1
|
selenium binding protein 1 |
| chr14_+_73790105 | 3.61 |
ENSMUST00000160507.8
ENSMUST00000022706.7 |
Sucla2
|
succinate-Coenzyme A ligase, ADP-forming, beta subunit |
| chr4_+_115458172 | 3.59 |
ENSMUST00000084342.6
|
Cyp4a32
|
cytochrome P450, family 4, subfamily a, polypeptide 32 |
| chr10_+_41395410 | 3.59 |
ENSMUST00000019962.15
|
Cd164
|
CD164 antigen |
| chr16_+_90017634 | 3.58 |
ENSMUST00000023707.11
|
Sod1
|
superoxide dismutase 1, soluble |
| chr18_+_51250748 | 3.57 |
ENSMUST00000116639.4
|
Prr16
|
proline rich 16 |
| chr16_-_97763780 | 3.57 |
ENSMUST00000232187.2
ENSMUST00000231263.2 ENSMUST00000052089.9 ENSMUST00000063605.15 ENSMUST00000113734.9 ENSMUST00000231560.2 ENSMUST00000232165.2 |
Zbtb21
C2cd2
|
zinc finger and BTB domain containing 21 C2 calcium-dependent domain containing 2 |
| chr7_-_103113358 | 3.56 |
ENSMUST00000214347.2
|
Olfr607
|
olfactory receptor 607 |
| chr1_-_155688551 | 3.50 |
ENSMUST00000194632.2
ENSMUST00000111764.8 |
Qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr1_-_155688635 | 3.49 |
ENSMUST00000035325.15
|
Qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr18_+_84869883 | 3.49 |
ENSMUST00000163083.2
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr8_-_45811774 | 3.48 |
ENSMUST00000155230.2
ENSMUST00000135912.8 |
Fam149a
|
family with sequence similarity 149, member A |
| chr19_-_7943365 | 3.47 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr5_-_83502966 | 3.44 |
ENSMUST00000053543.11
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chr1_+_175459559 | 3.43 |
ENSMUST00000040250.15
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr1_+_175459735 | 3.43 |
ENSMUST00000097458.4
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr1_-_90897329 | 3.41 |
ENSMUST00000130042.2
ENSMUST00000027529.12 |
Rab17
|
RAB17, member RAS oncogene family |
| chr15_-_89361571 | 3.37 |
ENSMUST00000165199.8
|
Arsa
|
arylsulfatase A |
| chr18_-_3281089 | 3.36 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr3_+_20039775 | 3.34 |
ENSMUST00000172860.2
|
Cp
|
ceruloplasmin |
| chr4_+_133246274 | 3.33 |
ENSMUST00000149807.2
ENSMUST00000042919.16 ENSMUST00000153811.2 ENSMUST00000105901.2 ENSMUST00000121797.2 |
Kdf1
|
keratinocyte differentiation factor 1 |
| chr5_+_31011140 | 3.31 |
ENSMUST00000202501.2
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr5_+_90666791 | 3.30 |
ENSMUST00000113179.9
ENSMUST00000128740.2 |
Afm
|
afamin |
| chr9_+_18848418 | 3.27 |
ENSMUST00000218385.2
|
Olfr832
|
olfactory receptor 832 |
| chr5_-_87686048 | 3.26 |
ENSMUST00000031199.11
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr1_+_107517726 | 3.24 |
ENSMUST00000000514.11
ENSMUST00000112706.4 |
Serpinb8
|
serine (or cysteine) peptidase inhibitor, clade B, member 8 |
| chr19_-_10807285 | 3.24 |
ENSMUST00000039043.15
|
Cd6
|
CD6 antigen |
| chr8_-_80021556 | 3.23 |
ENSMUST00000048718.4
|
Mmaa
|
methylmalonic aciduria (cobalamin deficiency) type A |
| chr19_-_39801188 | 3.21 |
ENSMUST00000162507.2
ENSMUST00000160476.9 ENSMUST00000239028.2 |
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40 |
| chr5_-_5564730 | 3.20 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr19_-_6885657 | 3.20 |
ENSMUST00000149261.8
|
Prdx5
|
peroxiredoxin 5 |
| chr9_+_7445822 | 3.19 |
ENSMUST00000034497.8
|
Mmp3
|
matrix metallopeptidase 3 |
| chr11_-_59937302 | 3.18 |
ENSMUST00000000310.14
ENSMUST00000102693.9 ENSMUST00000148512.2 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr19_-_58444336 | 3.17 |
ENSMUST00000131877.2
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chrM_+_10167 | 3.16 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr11_+_101556367 | 3.15 |
ENSMUST00000039388.3
|
Arl4d
|
ADP-ribosylation factor-like 4D |
| chr1_+_87983189 | 3.13 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou1f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.3 | 72.9 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 12.3 | 37.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 11.6 | 34.8 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 8.8 | 26.3 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 8.6 | 34.3 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 8.2 | 24.5 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 7.3 | 22.0 | GO:0009095 | tyrosine biosynthetic process(GO:0006571) aromatic amino acid family biosynthetic process(GO:0009073) aromatic amino acid family biosynthetic process, prephenate pathway(GO:0009095) |
| 6.9 | 75.5 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 6.1 | 24.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 6.0 | 18.0 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 5.9 | 82.9 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 5.7 | 187.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 5.2 | 31.3 | GO:0008355 | olfactory learning(GO:0008355) |
| 5.0 | 24.9 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 4.6 | 13.9 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 3.2 | 9.6 | GO:0034378 | chylomicron assembly(GO:0034378) regulation of chylomicron remnant clearance(GO:0090320) |
| 2.8 | 25.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 2.7 | 8.2 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 2.5 | 15.1 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 2.5 | 17.5 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 2.3 | 9.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.8 | 7.3 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 1.6 | 24.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.5 | 4.5 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 1.4 | 10.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 1.3 | 5.4 | GO:1904700 | granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 1.2 | 3.7 | GO:1904736 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 1.2 | 4.9 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 1.2 | 3.6 | GO:0042853 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) |
| 1.2 | 4.8 | GO:1903630 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 1.1 | 6.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 1.1 | 3.4 | GO:0002414 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 1.1 | 2.3 | GO:0009804 | coumarin metabolic process(GO:0009804) coumarin catabolic process(GO:0046226) |
| 1.1 | 3.3 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 1.1 | 3.3 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 1.1 | 5.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 1.0 | 6.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 1.0 | 6.1 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 1.0 | 1.0 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.9 | 26.3 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.9 | 2.8 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.9 | 3.5 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.9 | 4.4 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.8 | 4.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.8 | 3.3 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.8 | 3.2 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.8 | 4.7 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.8 | 2.3 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.8 | 3.8 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.7 | 2.8 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.7 | 4.8 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.7 | 2.6 | GO:1904000 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) positive regulation of eating behavior(GO:1904000) positive regulation of small intestine smooth muscle contraction(GO:1904349) negative regulation of energy homeostasis(GO:2000506) |
| 0.6 | 4.5 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 0.6 | 1.9 | GO:1904446 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) positive regulation of establishment of Sertoli cell barrier(GO:1904446) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.6 | 13.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.6 | 1.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.6 | 9.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.6 | 3.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.6 | 3.0 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.6 | 1.8 | GO:1904154 | trimming of terminal mannose on B branch(GO:0036509) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.6 | 4.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.6 | 1.7 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.6 | 2.3 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.6 | 2.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.5 | 2.7 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.5 | 4.8 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.5 | 3.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.5 | 14.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.5 | 0.5 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.5 | 1.0 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.5 | 3.0 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.5 | 1.5 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.5 | 5.4 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.5 | 2.4 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.5 | 5.8 | GO:0007567 | parturition(GO:0007567) |
| 0.5 | 1.4 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.5 | 6.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.5 | 4.6 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.5 | 39.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.5 | 3.6 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.5 | 1.4 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.4 | 1.3 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.4 | 2.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.4 | 4.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 1.3 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 0.4 | 1.7 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.4 | 2.1 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.4 | 7.4 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.4 | 6.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.4 | 2.5 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.4 | 6.2 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.4 | 4.9 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 | 4.3 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.4 | 4.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.4 | 1.2 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.4 | 31.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.4 | 1.5 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.4 | 4.8 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.4 | 8.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 1.1 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.4 | 1.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.4 | 3.2 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.3 | 2.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.3 | 1.4 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.3 | 2.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 2.3 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.3 | 3.0 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 1.3 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.3 | 3.5 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.3 | 6.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.3 | 0.9 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.3 | 3.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 2.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.3 | 1.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.3 | 1.2 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.3 | 0.9 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.3 | 0.9 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.3 | 10.8 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.3 | 1.4 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.3 | 1.4 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.3 | 7.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.3 | 1.7 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.3 | 0.8 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.3 | 5.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 0.3 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.3 | 1.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.3 | 3.4 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.3 | 3.6 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.3 | 1.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.3 | 1.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.3 | 2.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 | 2.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 1.3 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.2 | 1.7 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.2 | 1.7 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) |
| 0.2 | 0.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 0.7 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 1.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.2 | 0.9 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 5.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 1.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 6.9 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 0.6 | GO:0035801 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) metanephric comma-shaped body morphogenesis(GO:0072278) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.2 | 14.8 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.2 | 1.0 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 3.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.2 | 1.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 1.4 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.2 | 5.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 0.8 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.2 | 0.8 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.2 | 2.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.2 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 0.6 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.2 | 0.6 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.2 | 1.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 1.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.2 | 1.4 | GO:0045714 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.2 | 5.3 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.2 | 1.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 2.3 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.2 | 6.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 0.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 0.5 | GO:2000451 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.2 | 1.6 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 | 1.0 | GO:0009624 | response to nematode(GO:0009624) |
| 0.2 | 0.8 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.2 | 3.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 0.8 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.2 | 0.6 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.2 | 1.2 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 3.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 1.7 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.2 | 0.9 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 4.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 1.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 2.9 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 1.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 4.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.8 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.8 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.4 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 0.6 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.1 | 1.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.7 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.1 | 14.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 7.6 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.1 | 0.7 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 0.7 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.5 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 4.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 5.0 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 1.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 2.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 1.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.5 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 0.9 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 23.5 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.1 | 0.5 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 1.8 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 1.0 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.4 | GO:0048377 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.1 | 2.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.4 | GO:0010513 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 5.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 4.5 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 1.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.7 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 2.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.8 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 3.5 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 1.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.4 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 1.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 2.6 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 6.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 4.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 1.9 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.1 | 1.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 1.7 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 6.3 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 2.3 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 2.9 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.1 | 2.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.6 | GO:0060054 | monocyte extravasation(GO:0035696) positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 1.1 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 | 0.6 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 1.9 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.3 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.1 | 0.4 | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.1 | 5.2 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 1.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.1 | 1.0 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 1.8 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 1.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 1.4 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.1 | 0.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 1.0 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 2.6 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.1 | 2.8 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 0.5 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 1.0 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.4 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 1.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 0.8 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.9 | GO:0061525 | hindgut development(GO:0061525) |
| 0.1 | 1.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 4.5 | GO:0030258 | lipid modification(GO:0030258) |
| 0.1 | 0.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.2 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 1.3 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.1 | 1.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.6 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.7 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 1.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 1.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.8 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.7 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.4 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.1 | 0.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.2 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.2 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 2.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 7.0 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.1 | 1.9 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.7 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.1 | 3.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.2 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 0.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.8 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 | 1.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.7 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.5 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 34.5 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 1.0 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.8 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 3.4 | GO:1990267 | response to transition metal nanoparticle(GO:1990267) |
| 0.0 | 0.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 1.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 1.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.0 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 1.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.4 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.5 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 1.8 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 1.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 4.2 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 4.4 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 1.4 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.7 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.4 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.2 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.5 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.9 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.9 | GO:0006760 | folic acid-containing compound metabolic process(GO:0006760) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0070894 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 1.3 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 2.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 1.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 6.8 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 0.4 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.4 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0032329 | serine transport(GO:0032329) |
| 0.0 | 0.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0032305 | positive regulation of icosanoid secretion(GO:0032305) positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.3 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.8 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.9 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 2.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.3 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 2.3 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.0 | 1.4 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.3 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.0 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.2 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 1.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 52.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.4 | 12.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 2.1 | 8.2 | GO:0097637 | platelet dense granule membrane(GO:0031088) intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 1.3 | 10.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.9 | 7.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.9 | 2.7 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.8 | 11.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.7 | 2.9 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.7 | 5.6 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.7 | 7.4 | GO:0045179 | apical cortex(GO:0045179) |
| 0.6 | 1.9 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.5 | 5.3 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.5 | 13.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.5 | 1.4 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.4 | 2.9 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.4 | 1.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.4 | 2.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 32.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.3 | 31.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.3 | 1.3 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.3 | 1.3 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.3 | 3.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.3 | 32.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 2.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.3 | 0.9 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.3 | 0.9 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.3 | 1.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.3 | 1.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 3.2 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.3 | 4.8 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.3 | 1.8 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.3 | 1.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 0.7 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 1.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 0.9 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.2 | 3.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 1.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.2 | 0.8 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.2 | 2.8 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.2 | 0.8 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 8.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 3.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 3.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 3.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 1.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.2 | 4.6 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.2 | 4.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 13.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 3.0 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.2 | 12.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 1.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 0.7 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 5.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.5 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 0.7 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 0.8 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.2 | 5.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 0.7 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.2 | 333.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.1 | 15.8 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 1.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.4 | GO:0016014 | dystrobrevin complex(GO:0016014) |
| 0.1 | 1.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 6.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 3.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.7 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 2.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 3.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.6 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 3.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 3.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 21.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.9 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 16.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 4.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.7 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 1.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 3.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 4.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.2 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.9 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) filopodium tip(GO:0032433) RNA cap binding complex(GO:0034518) |
| 0.1 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.6 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.0 | 6.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 3.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0070531 | BRCA1-A complex(GO:0070531) BRISC complex(GO:0070552) |
| 0.0 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 1.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 8.2 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 5.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0005858 | axonemal dynein complex(GO:0005858) outer dynein arm(GO:0036157) |
| 0.0 | 5.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 7.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 2.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 6.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.4 | 52.2 | GO:0005009 | insulin-activated receptor activity(GO:0005009) |
| 11.6 | 34.8 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 11.2 | 44.8 | GO:0071614 | linoleic acid epoxygenase activity(GO:0071614) |
| 10.6 | 31.7 | GO:0005186 | pheromone activity(GO:0005186) |
| 9.3 | 55.5 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 8.2 | 24.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 7.1 | 21.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 6.2 | 24.9 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 6.2 | 254.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 4.4 | 17.5 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 4.0 | 12.1 | GO:0035731 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 3.9 | 115.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 3.7 | 22.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 3.5 | 28.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 3.5 | 13.9 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 3.3 | 13.4 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 3.3 | 13.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 1.9 | 15.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 1.8 | 7.4 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 1.7 | 6.9 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 1.6 | 26.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 1.6 | 4.7 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 1.4 | 7.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 1.4 | 4.1 | GO:0016034 | maleylacetoacetate isomerase activity(GO:0016034) |
| 1.3 | 9.4 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.3 | 21.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.2 | 6.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 1.2 | 3.6 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 1.2 | 3.6 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 1.2 | 4.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.1 | 3.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 1.1 | 4.4 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.1 | 21.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 1.1 | 8.6 | GO:0005534 | galactose binding(GO:0005534) |
| 1.0 | 6.1 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 1.0 | 50.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 1.0 | 17.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.9 | 2.8 | GO:0019002 | GMP binding(GO:0019002) |
| 0.9 | 5.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.9 | 7.9 | GO:0016160 | amylase activity(GO:0016160) |
| 0.9 | 2.6 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.9 | 0.9 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 0.9 | 4.3 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.8 | 7.5 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.8 | 7.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.8 | 4.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.8 | 3.3 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.8 | 3.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.8 | 3.2 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.8 | 24.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.8 | 2.3 | GO:0001566 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.7 | 3.0 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.7 | 7.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.7 | 3.5 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.7 | 8.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.6 | 1.9 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.6 | 4.4 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.6 | 15.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.6 | 3.6 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.6 | 10.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.6 | 3.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.6 | 5.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.6 | 2.9 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.6 | 2.3 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.6 | 2.8 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.6 | 1.7 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.5 | 1.6 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.5 | 1.6 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.5 | 1.4 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.5 | 2.3 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.4 | 4.5 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.4 | 4.4 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.4 | 12.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 3.0 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.4 | 5.9 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.4 | 3.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.4 | 2.1 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.4 | 4.9 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 62.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 2.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.4 | 2.3 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.4 | 1.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.4 | 3.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 3.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 1.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 1.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.3 | 22.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.3 | 1.0 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.3 | 2.3 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.3 | 1.6 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.3 | 0.9 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.3 | 1.8 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.3 | 0.9 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.3 | 3.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 0.9 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.3 | 4.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 0.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.3 | 3.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.3 | 3.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 8.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.3 | 1.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.3 | 1.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.3 | 0.8 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.3 | 2.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 4.9 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.3 | 7.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 1.2 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.2 | 3.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.9 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.2 | 1.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 1.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 2.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 2.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 0.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 5.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 2.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.2 | 1.0 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.2 | 28.2 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.2 | 2.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 1.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.2 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.2 | 4.6 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.2 | 0.8 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 1.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.9 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 0.6 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.2 | 1.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 0.9 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.2 | 7.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 0.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 4.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.2 | 0.7 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.2 | 0.5 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.2 | 0.6 | GO:0044729 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.2 | 1.2 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 1.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 3.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.3 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.1 | 14.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 1.8 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 4.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 8.9 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 1.6 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.5 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.8 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.1 | 0.7 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 2.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.8 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 8.7 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 2.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.7 | GO:0019863 | IgE binding(GO:0019863) |
| 0.1 | 1.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 2.3 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 1.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 2.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.7 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 2.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.9 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.1 | 1.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 3.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.3 | GO:0052599 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 0.1 | 2.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 1.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.3 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 2.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.5 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 1.8 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 6.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 3.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 4.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 2.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 3.0 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 1.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.3 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 43.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.3 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 3.5 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 3.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.2 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 2.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 1.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.3 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 2.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 1.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 1.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 2.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 2.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 1.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 2.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 6.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.9 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 2.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 2.9 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.5 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 2.9 | GO:0051015 | actin filament binding(GO:0051015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 8.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 1.5 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 1.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 11.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 13.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 7.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 0.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.9 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 1.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 2.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 4.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 5.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 3.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 3.2 | 3.2 | REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 2.2 | 24.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 2.1 | 64.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.3 | 5.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.1 | 12.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.9 | 19.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.8 | 40.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.6 | 13.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.5 | 7.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 3.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.4 | 7.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 13.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.3 | 9.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.3 | 6.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.3 | 56.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.3 | 4.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 4.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 4.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 0.9 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.2 | 2.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 3.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 7.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 7.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 2.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 2.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 4.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 2.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 3.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.4 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.1 | 7.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.9 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 3.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 3.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 6.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 2.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 3.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.9 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.6 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.3 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |