Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
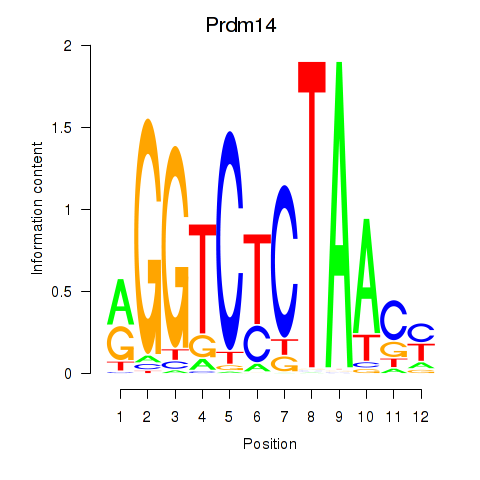
Results for Prdm14
Z-value: 0.42
Transcription factors associated with Prdm14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prdm14
|
ENSMUSG00000042414.8 | PR domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prdm14 | mm39_v1_chr1_-_13197387_13197473 | -0.37 | 2.7e-02 | Click! |
Activity profile of Prdm14 motif
Sorted Z-values of Prdm14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_88062508 | 2.15 |
ENSMUST00000113134.8
ENSMUST00000140092.8 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr4_+_133280680 | 2.13 |
ENSMUST00000042706.3
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr1_+_88030951 | 1.97 |
ENSMUST00000113135.6
ENSMUST00000113138.8 |
Ugt1a7c
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A7C UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr6_+_54016543 | 1.92 |
ENSMUST00000046856.14
|
Chn2
|
chimerin 2 |
| chr6_+_72332449 | 1.65 |
ENSMUST00000206064.2
|
Tmem150a
|
transmembrane protein 150A |
| chr17_-_35081129 | 1.40 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr6_+_72332423 | 1.33 |
ENSMUST00000069695.9
ENSMUST00000132243.3 |
Tmem150a
|
transmembrane protein 150A |
| chr17_-_35081456 | 1.32 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr6_+_91661074 | 1.16 |
ENSMUST00000205480.2
ENSMUST00000206545.2 |
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr8_+_46945826 | 1.12 |
ENSMUST00000110371.8
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr6_+_72333209 | 0.99 |
ENSMUST00000206531.2
|
Tmem150a
|
transmembrane protein 150A |
| chr17_+_44445659 | 0.99 |
ENSMUST00000239215.2
|
Clic5
|
chloride intracellular channel 5 |
| chr13_+_74787952 | 0.94 |
ENSMUST00000221822.2
ENSMUST00000221526.2 |
Erap1
|
endoplasmic reticulum aminopeptidase 1 |
| chr4_+_155646807 | 0.93 |
ENSMUST00000030939.14
|
Nadk
|
NAD kinase |
| chr9_+_94551929 | 0.90 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr15_-_76191301 | 0.85 |
ENSMUST00000171340.9
ENSMUST00000023222.13 ENSMUST00000164189.2 |
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr6_+_91661034 | 0.80 |
ENSMUST00000032185.9
|
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr14_-_72947422 | 0.70 |
ENSMUST00000089017.12
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chrX_+_67805443 | 0.68 |
ENSMUST00000069731.12
ENSMUST00000114647.8 |
Fmr1nb
|
Fmr1 neighbor |
| chr9_+_102595628 | 0.64 |
ENSMUST00000156485.2
ENSMUST00000145937.2 ENSMUST00000134483.2 ENSMUST00000190047.7 |
Amotl2
|
angiomotin-like 2 |
| chr9_+_65102635 | 0.52 |
ENSMUST00000216702.2
|
Parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr6_-_108162513 | 0.48 |
ENSMUST00000167338.8
ENSMUST00000172188.2 ENSMUST00000032191.16 |
Sumf1
|
sulfatase modifying factor 1 |
| chr6_+_87405968 | 0.47 |
ENSMUST00000032125.7
|
Bmp10
|
bone morphogenetic protein 10 |
| chr2_-_143853122 | 0.47 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr7_+_114344920 | 0.41 |
ENSMUST00000136645.8
ENSMUST00000169913.8 |
Insc
|
INSC spindle orientation adaptor protein |
| chr9_+_4376556 | 0.40 |
ENSMUST00000212075.2
|
Msantd4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chrX_+_98086187 | 0.38 |
ENSMUST00000036606.14
|
Stard8
|
START domain containing 8 |
| chr2_+_91560472 | 0.33 |
ENSMUST00000099712.10
ENSMUST00000111317.9 ENSMUST00000111316.9 ENSMUST00000045705.14 |
Ambra1
|
autophagy/beclin 1 regulator 1 |
| chr11_+_3599183 | 0.31 |
ENSMUST00000096441.5
|
Morc2a
|
microrchidia 2A |
| chrM_+_7779 | 0.31 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chrX_+_13147209 | 0.29 |
ENSMUST00000000804.7
|
Ddx3x
|
DEAD box helicase 3, X-linked |
| chr1_+_191709329 | 0.24 |
ENSMUST00000180463.3
ENSMUST00000181512.8 |
Rd3
|
retinal degeneration 3 |
| chrX_+_67805497 | 0.20 |
ENSMUST00000071848.7
|
Fmr1nb
|
Fmr1 neighbor |
| chr4_+_152358648 | 0.19 |
ENSMUST00000105650.8
ENSMUST00000105651.8 |
Gpr153
|
G protein-coupled receptor 153 |
| chr1_-_173703424 | 0.18 |
ENSMUST00000186442.7
|
Mndal
|
myeloid nuclear differentiation antigen like |
| chr11_+_43046476 | 0.16 |
ENSMUST00000238415.2
|
Atp10b
|
ATPase, class V, type 10B |
| chr2_+_128809268 | 0.14 |
ENSMUST00000110320.9
ENSMUST00000110319.3 |
Zc3h6
|
zinc finger CCCH type containing 6 |
| chr4_+_116983973 | 0.14 |
ENSMUST00000134074.8
|
Dynlt4
|
dynein light chain Tctex-type 4 |
| chr7_+_18721325 | 0.14 |
ENSMUST00000063563.9
|
Nanos2
|
nanos C2HC-type zinc finger 2 |
| chr9_-_14815228 | 0.08 |
ENSMUST00000034409.14
ENSMUST00000117620.8 |
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr7_+_27510238 | 0.07 |
ENSMUST00000080175.7
|
Zfp626
|
zinc finger protein 626 |
| chrX_-_93585668 | 0.06 |
ENSMUST00000026142.8
|
Maged1
|
MAGE family member D1 |
| chr15_+_99499252 | 0.06 |
ENSMUST00000230075.2
ENSMUST00000023754.6 |
Aqp6
|
aquaporin 6 |
| chr10_-_128579879 | 0.05 |
ENSMUST00000026414.9
|
Dgka
|
diacylglycerol kinase, alpha |
| chr4_+_128777339 | 0.05 |
ENSMUST00000035667.9
|
Trim62
|
tripartite motif-containing 62 |
| chr4_+_116983995 | 0.05 |
ENSMUST00000062206.3
|
Dynlt4
|
dynein light chain Tctex-type 4 |
| chr18_-_70663382 | 0.05 |
ENSMUST00000043286.15
|
Poli
|
polymerase (DNA directed), iota |
| chr15_-_73114855 | 0.04 |
ENSMUST00000227686.2
|
Ptk2
|
PTK2 protein tyrosine kinase 2 |
| chr4_-_133399909 | 0.03 |
ENSMUST00000062118.11
ENSMUST00000067902.13 |
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr4_+_119671688 | 0.03 |
ENSMUST00000106307.9
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr18_-_70663209 | 0.03 |
ENSMUST00000161542.8
ENSMUST00000159389.8 |
Poli
|
polymerase (DNA directed), iota |
| chr5_-_38842084 | 0.03 |
ENSMUST00000057258.11
ENSMUST00000178760.2 ENSMUST00000179555.8 ENSMUST00000180214.8 |
Zfp518b
|
zinc finger protein 518B |
| chr8_-_93774820 | 0.02 |
ENSMUST00000095211.5
|
Ces1a
|
carboxylesterase 1A |
| chr12_+_69939879 | 0.02 |
ENSMUST00000021466.10
|
Atl1
|
atlastin GTPase 1 |
| chr17_+_27000034 | 0.02 |
ENSMUST00000015725.16
ENSMUST00000135824.8 ENSMUST00000137989.2 |
Bnip1
|
BCL2/adenovirus E1B interacting protein 1 |
| chr9_-_14815050 | 0.01 |
ENSMUST00000148155.2
ENSMUST00000121116.8 |
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr15_-_73579236 | 0.01 |
ENSMUST00000064166.5
|
Gpr20
|
G protein-coupled receptor 20 |
| chr9_-_14815163 | 0.01 |
ENSMUST00000069408.10
|
Izumo1r
|
IZUMO1 receptor, JUNO |
| chr6_-_42437951 | 0.00 |
ENSMUST00000090156.2
|
Olfr458
|
olfactory receptor 458 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prdm14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.7 | 2.0 | GO:0015734 | beta-alanine transport(GO:0001762) taurine transport(GO:0015734) |
| 0.2 | 2.1 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 2.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.9 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 4.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.5 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.9 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.9 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.5 | GO:0010612 | regulation of cardiac muscle adaptation(GO:0010612) positive regulation of sarcomere organization(GO:0060298) regulation of cardiac muscle hypertrophy in response to stress(GO:1903242) |
| 0.0 | 1.0 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.7 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 2.1 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.1 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0005368 | beta-alanine transmembrane transporter activity(GO:0001761) taurine transmembrane transporter activity(GO:0005368) taurine:sodium symporter activity(GO:0005369) |
| 0.2 | 1.1 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.1 | 4.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.9 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.8 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 2.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 1.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |