Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Spib
Z-value: 4.04
Transcription factors associated with Spib
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Spib
|
ENSMUSG00000008193.14 | Spi-B transcription factor (Spi-1/PU.1 related) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Spib | mm39_v1_chr7_-_44181477_44181502 | 0.87 | 5.8e-12 | Click! |
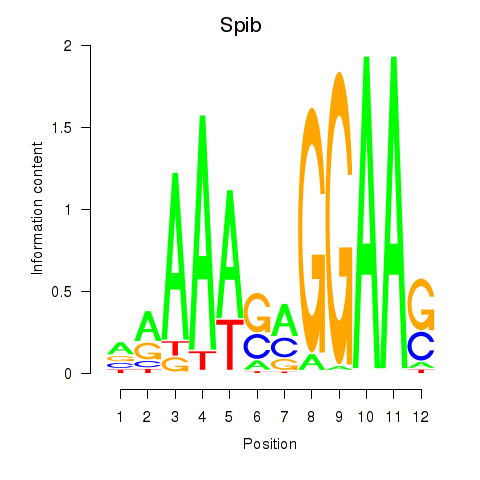
Activity profile of Spib motif
Sorted Z-values of Spib motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_136918671 | 41.29 |
ENSMUST00000032344.12
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr14_+_80237691 | 34.37 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr5_+_149201577 | 27.78 |
ENSMUST00000071130.5
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr11_-_103235475 | 23.58 |
ENSMUST00000041385.14
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr11_-_16958647 | 23.27 |
ENSMUST00000102881.10
|
Plek
|
pleckstrin |
| chr5_+_149202157 | 23.20 |
ENSMUST00000200806.4
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr5_-_134258435 | 23.06 |
ENSMUST00000016094.13
ENSMUST00000111275.8 ENSMUST00000144086.2 |
Ncf1
|
neutrophil cytosolic factor 1 |
| chr9_+_55997246 | 22.82 |
ENSMUST00000059206.8
|
Pstpip1
|
proline-serine-threonine phosphatase-interacting protein 1 |
| chr15_-_66684442 | 22.80 |
ENSMUST00000100572.10
|
Sla
|
src-like adaptor |
| chr7_+_99184645 | 20.94 |
ENSMUST00000098266.9
ENSMUST00000179755.8 |
Arrb1
|
arrestin, beta 1 |
| chr11_+_11635908 | 20.51 |
ENSMUST00000065433.12
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr7_-_44888220 | 20.27 |
ENSMUST00000210372.2
ENSMUST00000209779.2 ENSMUST00000098461.10 ENSMUST00000211373.2 |
Cd37
|
CD37 antigen |
| chr7_-_126303351 | 19.80 |
ENSMUST00000106364.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr6_-_136918495 | 19.69 |
ENSMUST00000111892.8
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr7_-_126303887 | 19.65 |
ENSMUST00000131415.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr15_+_78210190 | 19.24 |
ENSMUST00000229034.2
ENSMUST00000096355.4 |
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr7_-_45570828 | 19.07 |
ENSMUST00000038876.13
|
Emp3
|
epithelial membrane protein 3 |
| chr1_-_170886924 | 18.71 |
ENSMUST00000164044.8
ENSMUST00000169017.8 |
Fcgr3
|
Fc receptor, IgG, low affinity III |
| chr2_-_181333597 | 18.36 |
ENSMUST00000108778.8
ENSMUST00000165416.8 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr2_+_24235300 | 18.15 |
ENSMUST00000114485.9
ENSMUST00000114482.3 |
Il1rn
|
interleukin 1 receptor antagonist |
| chr14_-_70864448 | 18.14 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr15_+_78210242 | 17.95 |
ENSMUST00000229678.2
ENSMUST00000231888.2 |
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr17_+_57586094 | 17.38 |
ENSMUST00000169220.9
ENSMUST00000005889.16 ENSMUST00000112870.5 |
Vav1
|
vav 1 oncogene |
| chr15_-_63932176 | 17.08 |
ENSMUST00000226675.2
ENSMUST00000228226.2 ENSMUST00000227024.2 |
Cyrib
|
CYFIP related Rac1 interactor B |
| chr15_-_78189917 | 16.87 |
ENSMUST00000096356.5
|
Csf2rb2
|
colony stimulating factor 2 receptor, beta 2, low-affinity (granulocyte-macrophage) |
| chr2_+_22664094 | 16.83 |
ENSMUST00000014290.15
|
Apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr11_-_34674677 | 16.71 |
ENSMUST00000093193.12
ENSMUST00000101365.9 |
Dock2
|
dedicator of cyto-kinesis 2 |
| chr7_-_45570538 | 16.61 |
ENSMUST00000210297.2
|
Emp3
|
epithelial membrane protein 3 |
| chr4_+_130640611 | 16.58 |
ENSMUST00000156225.8
ENSMUST00000156742.8 |
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr15_-_36609208 | 16.22 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr2_-_165242307 | 16.16 |
ENSMUST00000029213.5
|
Ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr15_+_78209920 | 16.08 |
ENSMUST00000230264.3
|
Csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr9_-_114610879 | 15.94 |
ENSMUST00000084867.9
ENSMUST00000216760.2 ENSMUST00000035009.16 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr15_-_78456898 | 15.93 |
ENSMUST00000043214.8
|
Rac2
|
Rac family small GTPase 2 |
| chr1_-_170755136 | 15.91 |
ENSMUST00000046322.14
ENSMUST00000159171.2 |
Fcrla
|
Fc receptor-like A |
| chr6_+_38895902 | 15.73 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr14_-_70864666 | 15.40 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr4_+_140428777 | 15.31 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr15_-_82917495 | 15.19 |
ENSMUST00000231165.2
|
Nfam1
|
Nfat activating molecule with ITAM motif 1 |
| chr2_-_181335697 | 15.19 |
ENSMUST00000108779.8
ENSMUST00000108769.8 ENSMUST00000108772.8 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr4_+_130640436 | 15.09 |
ENSMUST00000151698.8
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr19_+_4204605 | 15.05 |
ENSMUST00000061086.9
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr7_-_45570674 | 14.95 |
ENSMUST00000210939.2
|
Emp3
|
epithelial membrane protein 3 |
| chr15_+_103362195 | 14.94 |
ENSMUST00000047405.9
|
Nckap1l
|
NCK associated protein 1 like |
| chr17_+_35268942 | 14.86 |
ENSMUST00000007257.10
|
Clic1
|
chloride intracellular channel 1 |
| chr6_+_129326927 | 14.75 |
ENSMUST00000065289.6
|
Clec12a
|
C-type lectin domain family 12, member a |
| chr13_-_113237505 | 14.53 |
ENSMUST00000224282.2
ENSMUST00000023897.7 |
Gzma
|
granzyme A |
| chr15_-_78189822 | 13.94 |
ENSMUST00000230115.3
|
Csf2rb2
|
colony stimulating factor 2 receptor, beta 2, low-affinity (granulocyte-macrophage) |
| chr7_-_126303689 | 13.92 |
ENSMUST00000135087.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr1_-_171061902 | 13.69 |
ENSMUST00000079957.12
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr7_-_44888532 | 13.65 |
ENSMUST00000033063.15
|
Cd37
|
CD37 antigen |
| chr13_+_20978283 | 13.13 |
ENSMUST00000021757.5
ENSMUST00000221982.2 |
Aoah
|
acyloxyacyl hydrolase |
| chr2_-_38895586 | 12.97 |
ENSMUST00000080861.6
|
Rpl35
|
ribosomal protein L35 |
| chr7_-_44888465 | 12.55 |
ENSMUST00000210078.2
|
Cd37
|
CD37 antigen |
| chr16_+_36755338 | 11.84 |
ENSMUST00000023531.15
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr19_+_6449887 | 11.82 |
ENSMUST00000146601.8
ENSMUST00000150713.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr2_-_181335767 | 11.73 |
ENSMUST00000002532.9
|
Rgs19
|
regulator of G-protein signaling 19 |
| chr2_-_181335518 | 11.68 |
ENSMUST00000108776.8
ENSMUST00000108771.2 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr1_+_87548026 | 11.37 |
ENSMUST00000169754.8
ENSMUST00000042275.15 ENSMUST00000168783.8 |
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr19_+_6135013 | 11.35 |
ENSMUST00000025704.3
|
Cdca5
|
cell division cycle associated 5 |
| chr7_-_3723381 | 10.87 |
ENSMUST00000078451.7
|
Pirb
|
paired Ig-like receptor B |
| chr12_-_28685849 | 10.84 |
ENSMUST00000221871.2
|
Rps7
|
ribosomal protein S7 |
| chr17_+_48539782 | 10.79 |
ENSMUST00000113251.10
ENSMUST00000048782.7 |
Trem1
|
triggering receptor expressed on myeloid cells 1 |
| chr2_+_91480460 | 10.78 |
ENSMUST00000111331.9
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr16_-_36486429 | 10.64 |
ENSMUST00000089620.11
|
Cd86
|
CD86 antigen |
| chr9_+_106080307 | 10.47 |
ENSMUST00000024047.12
ENSMUST00000216348.2 |
Twf2
|
twinfilin actin binding protein 2 |
| chr12_+_105996961 | 10.34 |
ENSMUST00000220629.2
|
Vrk1
|
vaccinia related kinase 1 |
| chr8_-_89362745 | 10.11 |
ENSMUST00000034087.9
|
Snx20
|
sorting nexin 20 |
| chr12_-_28685913 | 9.76 |
ENSMUST00000074267.5
|
Rps7
|
ribosomal protein S7 |
| chr7_+_3339077 | 9.56 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chrX_+_20570145 | 9.55 |
ENSMUST00000033383.3
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr3_-_59118293 | 9.38 |
ENSMUST00000040622.3
|
P2ry13
|
purinergic receptor P2Y, G-protein coupled 13 |
| chr7_+_3339059 | 9.32 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr12_-_79027531 | 9.31 |
ENSMUST00000174072.8
|
Tmem229b
|
transmembrane protein 229B |
| chr1_-_170755109 | 9.30 |
ENSMUST00000162136.2
ENSMUST00000162887.2 |
Fcrla
|
Fc receptor-like A |
| chr1_+_171509565 | 9.26 |
ENSMUST00000015499.14
ENSMUST00000068584.7 |
Cd48
|
CD48 antigen |
| chr2_+_90927053 | 9.26 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr4_+_119052476 | 9.19 |
ENSMUST00000030395.9
|
Svbp
|
small vasohibin binding protein |
| chr19_+_10582920 | 9.18 |
ENSMUST00000236280.2
|
Ddb1
|
damage specific DNA binding protein 1 |
| chrX_+_141464722 | 9.07 |
ENSMUST00000112896.9
|
Tmem164
|
transmembrane protein 164 |
| chr2_+_91480513 | 9.04 |
ENSMUST00000090614.11
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr11_+_46701619 | 8.94 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr10_-_81360059 | 8.89 |
ENSMUST00000043709.8
|
Gna15
|
guanine nucleotide binding protein, alpha 15 |
| chr4_-_115980813 | 8.77 |
ENSMUST00000102704.4
ENSMUST00000102705.10 |
Rad54l
|
RAD54 like (S. cerevisiae) |
| chr17_+_34823236 | 8.68 |
ENSMUST00000174041.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr3_-_96201248 | 8.58 |
ENSMUST00000029748.8
|
Fcgr1
|
Fc receptor, IgG, high affinity I |
| chr7_+_126808016 | 8.57 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr4_+_109200225 | 8.48 |
ENSMUST00000030281.12
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr11_+_117006020 | 8.45 |
ENSMUST00000103026.10
ENSMUST00000090433.6 |
Sec14l1
|
SEC14-like lipid binding 1 |
| chr7_-_99132843 | 8.43 |
ENSMUST00000208532.2
ENSMUST00000107096.2 ENSMUST00000032998.13 |
Rps3
|
ribosomal protein S3 |
| chr18_+_75133519 | 8.37 |
ENSMUST00000079716.6
|
Rpl17
|
ribosomal protein L17 |
| chr6_-_83302890 | 8.33 |
ENSMUST00000204472.2
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| chr5_-_116162415 | 8.07 |
ENSMUST00000031486.14
ENSMUST00000111999.8 |
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr7_-_28931873 | 8.06 |
ENSMUST00000085818.6
|
Kcnk6
|
potassium inwardly-rectifying channel, subfamily K, member 6 |
| chr9_+_21437440 | 8.04 |
ENSMUST00000086361.12
ENSMUST00000173769.3 |
AB124611
|
cDNA sequence AB124611 |
| chr2_+_24276616 | 8.02 |
ENSMUST00000166388.2
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr10_+_81229443 | 7.98 |
ENSMUST00000118206.2
|
Smim24
|
small integral membrane protein 24 |
| chr3_-_105839980 | 7.93 |
ENSMUST00000098758.5
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr2_+_120294046 | 7.76 |
ENSMUST00000028749.15
ENSMUST00000110721.9 ENSMUST00000239364.2 |
Capn3
|
calpain 3 |
| chr7_+_43057611 | 7.76 |
ENSMUST00000005592.7
|
Siglecg
|
sialic acid binding Ig-like lectin G |
| chr13_+_30933209 | 7.72 |
ENSMUST00000021784.10
ENSMUST00000110307.3 ENSMUST00000222125.2 |
Irf4
|
interferon regulatory factor 4 |
| chr11_+_87628356 | 7.40 |
ENSMUST00000093955.12
|
Supt4a
|
SPT4A, DSIF elongation factor subunit |
| chr19_+_6449776 | 7.36 |
ENSMUST00000113468.8
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr4_+_119052693 | 7.36 |
ENSMUST00000097908.4
|
Svbp
|
small vasohibin binding protein |
| chr19_+_10582987 | 7.24 |
ENSMUST00000237337.2
|
Ddb1
|
damage specific DNA binding protein 1 |
| chr17_+_8144822 | 7.18 |
ENSMUST00000036370.8
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr10_+_81093110 | 7.13 |
ENSMUST00000117488.8
ENSMUST00000105328.10 ENSMUST00000121205.8 |
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr7_-_100505486 | 7.07 |
ENSMUST00000139604.8
|
Relt
|
RELT tumor necrosis factor receptor |
| chr1_+_171594690 | 6.88 |
ENSMUST00000015460.5
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr11_-_115024807 | 6.84 |
ENSMUST00000106561.8
ENSMUST00000051264.14 ENSMUST00000106562.3 |
Cd300lf
|
CD300 molecule like family member F |
| chr17_-_35827676 | 6.78 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr17_+_36179273 | 6.75 |
ENSMUST00000190496.2
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr9_-_114811807 | 6.70 |
ENSMUST00000053150.8
|
Rps27rt
|
ribosomal protein S27, retrogene |
| chr13_+_104365432 | 6.69 |
ENSMUST00000070761.10
ENSMUST00000225557.2 |
Cenpk
|
centromere protein K |
| chr11_-_97673203 | 6.68 |
ENSMUST00000128801.2
ENSMUST00000103146.5 |
Rpl23
|
ribosomal protein L23 |
| chr10_-_88192852 | 6.54 |
ENSMUST00000020249.2
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr2_+_25313240 | 6.42 |
ENSMUST00000134259.8
ENSMUST00000100320.5 |
Fut7
|
fucosyltransferase 7 |
| chr9_+_64718596 | 6.40 |
ENSMUST00000038890.6
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr4_+_119052548 | 6.32 |
ENSMUST00000106345.3
|
Svbp
|
small vasohibin binding protein |
| chr4_+_132903646 | 6.27 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr7_-_80994933 | 6.15 |
ENSMUST00000080813.5
|
Rps17
|
ribosomal protein S17 |
| chr8_+_72943455 | 6.11 |
ENSMUST00000072097.14
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr2_+_32611067 | 6.11 |
ENSMUST00000074248.11
|
Sh2d3c
|
SH2 domain containing 3C |
| chr11_+_70021155 | 6.10 |
ENSMUST00000041550.12
ENSMUST00000165951.8 |
Mgl2
|
macrophage galactose N-acetyl-galactosamine specific lectin 2 |
| chr6_-_124710030 | 6.09 |
ENSMUST00000173647.2
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr2_+_163916042 | 5.91 |
ENSMUST00000018353.14
|
Stk4
|
serine/threonine kinase 4 |
| chr13_-_13568106 | 5.81 |
ENSMUST00000021738.10
ENSMUST00000220628.2 |
Gpr137b
|
G protein-coupled receptor 137B |
| chr4_-_136620376 | 5.80 |
ENSMUST00000046332.6
|
C1qc
|
complement component 1, q subcomponent, C chain |
| chr12_+_98234884 | 5.74 |
ENSMUST00000075072.6
|
Gpr65
|
G-protein coupled receptor 65 |
| chr6_+_122929591 | 5.69 |
ENSMUST00000088468.7
|
Clec4a3
|
C-type lectin domain family 4, member a3 |
| chr4_-_136613498 | 5.68 |
ENSMUST00000046384.9
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide |
| chr8_-_25592385 | 5.60 |
ENSMUST00000064883.14
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr19_+_6108240 | 5.38 |
ENSMUST00000237840.2
|
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) |
| chr10_-_40178182 | 5.38 |
ENSMUST00000099945.6
ENSMUST00000238953.2 ENSMUST00000238969.2 |
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr4_-_117232650 | 5.30 |
ENSMUST00000094853.9
|
Rnf220
|
ring finger protein 220 |
| chr4_-_141602190 | 5.30 |
ENSMUST00000036854.4
|
Efhd2
|
EF hand domain containing 2 |
| chr1_-_125362321 | 5.25 |
ENSMUST00000191544.7
|
Actr3
|
ARP3 actin-related protein 3 |
| chr15_+_97259060 | 5.23 |
ENSMUST00000228521.2
ENSMUST00000226495.2 |
Pced1b
|
PC-esterase domain containing 1B |
| chr8_-_123302187 | 5.08 |
ENSMUST00000213062.2
|
Aprt
|
adenine phosphoribosyl transferase |
| chrX_+_41238193 | 5.08 |
ENSMUST00000115073.9
ENSMUST00000115072.8 |
Stag2
|
stromal antigen 2 |
| chr19_+_6107874 | 5.02 |
ENSMUST00000178310.9
|
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) |
| chr6_-_124710084 | 4.97 |
ENSMUST00000112484.10
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr15_+_80832685 | 4.96 |
ENSMUST00000023043.10
ENSMUST00000164806.6 ENSMUST00000207170.2 ENSMUST00000168756.8 |
Adsl
|
adenylosuccinate lyase |
| chr19_+_6107957 | 4.89 |
ENSMUST00000237859.2
ENSMUST00000043074.14 ENSMUST00000236336.2 ENSMUST00000179142.2 ENSMUST00000236217.2 |
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) |
| chr7_-_3848050 | 4.85 |
ENSMUST00000108615.10
ENSMUST00000119469.2 |
Pira2
|
paired-Ig-like receptor A2 |
| chr7_-_126807581 | 4.83 |
ENSMUST00000120705.3
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr2_+_43638814 | 4.81 |
ENSMUST00000112824.8
ENSMUST00000055776.8 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chr13_-_23650045 | 4.75 |
ENSMUST00000041674.14
ENSMUST00000110434.2 |
Btn1a1
|
butyrophilin, subfamily 1, member A1 |
| chr14_+_75368939 | 4.61 |
ENSMUST00000125833.8
ENSMUST00000124499.8 |
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr8_+_79754980 | 4.55 |
ENSMUST00000087927.11
ENSMUST00000098614.9 |
Zfp827
|
zinc finger protein 827 |
| chr8_+_121215155 | 4.37 |
ENSMUST00000034279.16
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr19_-_11313471 | 4.34 |
ENSMUST00000056035.9
ENSMUST00000067532.11 |
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr1_-_38704028 | 4.29 |
ENSMUST00000039827.14
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr4_+_124608569 | 4.27 |
ENSMUST00000030734.5
|
Sf3a3
|
splicing factor 3a, subunit 3 |
| chr8_+_95472218 | 4.24 |
ENSMUST00000034231.4
|
Ccl22
|
chemokine (C-C motif) ligand 22 |
| chr11_+_120348919 | 4.10 |
ENSMUST00000058370.14
ENSMUST00000175970.8 ENSMUST00000176120.2 |
Ccdc137
|
coiled-coil domain containing 137 |
| chr6_+_60921456 | 4.09 |
ENSMUST00000129603.4
ENSMUST00000204333.2 |
Mmrn1
|
multimerin 1 |
| chr4_-_135780660 | 4.03 |
ENSMUST00000102536.11
|
Rpl11
|
ribosomal protein L11 |
| chr3_-_89959770 | 4.02 |
ENSMUST00000029553.16
ENSMUST00000195995.5 ENSMUST00000064639.15 ENSMUST00000199834.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr10_-_6930376 | 3.92 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr14_-_66071412 | 3.86 |
ENSMUST00000022613.10
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr11_-_115590318 | 3.85 |
ENSMUST00000106497.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chr7_+_101520843 | 3.84 |
ENSMUST00000210984.2
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chrX_+_7656225 | 3.79 |
ENSMUST00000136930.8
ENSMUST00000115675.9 ENSMUST00000101694.10 |
Gripap1
|
GRIP1 associated protein 1 |
| chr7_+_127078371 | 3.75 |
ENSMUST00000205432.3
|
Fbrs
|
fibrosin |
| chr6_-_120799500 | 3.67 |
ENSMUST00000204699.2
|
Atp6v1e1
|
ATPase, H+ transporting, lysosomal V1 subunit E1 |
| chr3_-_89959739 | 3.60 |
ENSMUST00000199929.2
ENSMUST00000090908.11 ENSMUST00000198322.5 ENSMUST00000196843.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr5_+_86219593 | 3.60 |
ENSMUST00000198435.5
ENSMUST00000031171.9 |
Stap1
|
signal transducing adaptor family member 1 |
| chr2_-_91480096 | 3.52 |
ENSMUST00000099714.10
ENSMUST00000111333.2 |
Zfp408
|
zinc finger protein 408 |
| chr19_-_40982576 | 3.50 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr8_-_25730878 | 3.44 |
ENSMUST00000210488.2
ENSMUST00000210933.2 |
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr3_-_135313982 | 3.44 |
ENSMUST00000132668.8
|
Nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
| chr18_-_43610829 | 3.43 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr7_-_46658144 | 3.42 |
ENSMUST00000061639.10
|
Spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr17_-_50497682 | 3.30 |
ENSMUST00000044503.14
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr9_+_123902143 | 3.30 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr19_-_5711650 | 3.26 |
ENSMUST00000236006.2
|
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr6_+_83303052 | 3.26 |
ENSMUST00000038658.15
|
Mob1a
|
MOB kinase activator 1A |
| chr14_-_65830463 | 3.23 |
ENSMUST00000225355.2
ENSMUST00000022609.7 |
Elp3
|
elongator acetyltransferase complex subunit 3 |
| chr4_-_92079986 | 3.20 |
ENSMUST00000123179.2
|
Gm12666
|
predicted gene 12666 |
| chr9_+_64718708 | 3.18 |
ENSMUST00000213926.2
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr2_-_60793536 | 3.16 |
ENSMUST00000028347.13
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr3_+_60380463 | 3.15 |
ENSMUST00000195077.6
ENSMUST00000193647.6 ENSMUST00000195001.2 ENSMUST00000192807.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr7_+_127187910 | 3.09 |
ENSMUST00000205694.2
ENSMUST00000033088.8 ENSMUST00000206914.2 |
Rnf40
|
ring finger protein 40 |
| chr14_-_30645711 | 3.08 |
ENSMUST00000006697.17
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr5_+_115680964 | 3.05 |
ENSMUST00000137716.8
|
Pxn
|
paxillin |
| chr11_+_113548201 | 3.04 |
ENSMUST00000148736.8
ENSMUST00000142069.8 ENSMUST00000134418.8 |
Cog1
|
component of oligomeric golgi complex 1 |
| chr16_-_89757693 | 2.98 |
ENSMUST00000002588.11
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr10_+_96453408 | 2.93 |
ENSMUST00000218953.2
|
Btg1
|
BTG anti-proliferation factor 1 |
| chrX_-_93166992 | 2.92 |
ENSMUST00000088102.12
ENSMUST00000113927.8 |
Zfx
|
zinc finger protein X-linked |
| chr5_-_140368482 | 2.90 |
ENSMUST00000196566.5
|
Snx8
|
sorting nexin 8 |
| chr13_+_102830029 | 2.90 |
ENSMUST00000022124.10
ENSMUST00000171267.2 ENSMUST00000167144.2 ENSMUST00000170878.2 |
Cd180
|
CD180 antigen |
| chr1_+_75498162 | 2.90 |
ENSMUST00000027414.16
ENSMUST00000113553.2 |
Stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr6_+_83302998 | 2.89 |
ENSMUST00000055261.11
|
Mob1a
|
MOB kinase activator 1A |
| chr5_+_123280250 | 2.83 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr9_+_106099797 | 2.80 |
ENSMUST00000062241.11
|
Tlr9
|
toll-like receptor 9 |
| chr11_-_115590133 | 2.78 |
ENSMUST00000106499.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chr12_-_32258331 | 2.76 |
ENSMUST00000220366.2
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr2_-_91795910 | 2.74 |
ENSMUST00000239257.2
|
Dgkz
|
diacylglycerol kinase zeta |
| chrX_+_168468186 | 2.74 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Spib
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.1 | 27.3 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 8.5 | 51.0 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 7.9 | 23.6 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 7.8 | 23.3 | GO:0060305 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 7.7 | 23.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 6.9 | 55.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 6.8 | 20.5 | GO:0045660 | positive regulation of neutrophil differentiation(GO:0045660) |
| 6.8 | 61.0 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 5.6 | 33.5 | GO:0070560 | calcium-mediated signaling using extracellular calcium source(GO:0035585) protein secretion by platelet(GO:0070560) |
| 5.1 | 15.3 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 4.6 | 13.7 | GO:0001805 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 4.3 | 47.3 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 4.0 | 16.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 3.7 | 14.9 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 3.6 | 18.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 3.4 | 10.3 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 3.3 | 19.8 | GO:0032439 | endosome localization(GO:0032439) negative regulation of vacuolar transport(GO:1903336) |
| 2.8 | 11.4 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 2.7 | 16.4 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 2.6 | 20.9 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 2.6 | 7.7 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 2.4 | 9.8 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 2.4 | 16.8 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 2.4 | 18.9 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 2.3 | 16.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 2.3 | 11.4 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 2.1 | 8.4 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 2.0 | 31.3 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 1.9 | 7.8 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 1.9 | 9.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 1.8 | 55.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 1.8 | 3.6 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 1.8 | 5.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 1.7 | 6.8 | GO:0035963 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) interleukin-4-mediated signaling pathway(GO:0035771) response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) negative regulation of apoptotic cell clearance(GO:2000426) |
| 1.6 | 15.9 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 1.5 | 10.8 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 1.5 | 15.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 1.5 | 5.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.5 | 14.5 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 1.4 | 11.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 1.3 | 8.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 1.3 | 5.1 | GO:0046083 | adenine salvage(GO:0006168) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 1.2 | 5.0 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 1.2 | 8.5 | GO:0015871 | choline transport(GO:0015871) |
| 1.1 | 3.4 | GO:1904629 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) response to diterpene(GO:1904629) cellular response to diterpene(GO:1904630) response to glucoside(GO:1904631) cellular response to glucoside(GO:1904632) |
| 1.1 | 4.6 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 1.1 | 8.9 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 1.0 | 10.5 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 1.0 | 15.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 1.0 | 6.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 1.0 | 13.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 1.0 | 3.0 | GO:1990751 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 1.0 | 3.0 | GO:1903977 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of glial cell migration(GO:1903977) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.9 | 2.8 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.9 | 7.5 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.9 | 10.7 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.9 | 8.8 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.9 | 15.7 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.9 | 3.4 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.8 | 8.5 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.7 | 5.8 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) negative regulation of macrophage differentiation(GO:0045650) |
| 0.7 | 6.2 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.7 | 3.3 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.6 | 2.6 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.6 | 3.7 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.6 | 7.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.6 | 13.9 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.6 | 36.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.6 | 1.8 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.6 | 1.7 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.6 | 8.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.5 | 2.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.5 | 1.6 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.5 | 2.1 | GO:0033379 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.5 | 1.0 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.5 | 1.5 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.5 | 1.5 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.4 | 18.0 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.4 | 1.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.4 | 2.4 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.4 | 8.8 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.4 | 2.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 6.6 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.3 | 21.8 | GO:0010596 | negative regulation of endothelial cell migration(GO:0010596) |
| 0.3 | 7.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.3 | 2.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 9.4 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.3 | 5.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.3 | 6.3 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.3 | 1.8 | GO:2000354 | negative regulation of immature T cell proliferation(GO:0033087) regulation of ovarian follicle development(GO:2000354) |
| 0.3 | 3.8 | GO:0070572 | positive regulation of neuron projection regeneration(GO:0070572) |
| 0.3 | 27.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.3 | 8.3 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.3 | 3.0 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 3.8 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.3 | 2.5 | GO:0060762 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) germline stem cell asymmetric division(GO:0098728) |
| 0.3 | 0.8 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.3 | 4.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.3 | 2.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 19.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.3 | 9.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.9 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 1.2 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.2 | 2.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 2.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 4.6 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.2 | 1.5 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.2 | 6.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 6.8 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.2 | 14.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 1.0 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 2.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 72.2 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.2 | 1.4 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.2 | 5.7 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.2 | 0.5 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.6 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 2.0 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 4.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 3.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 2.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 2.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 23.2 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.1 | 6.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 2.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.5 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 23.8 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 3.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 13.1 | GO:0044264 | cellular polysaccharide metabolic process(GO:0044264) |
| 0.1 | 2.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 2.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 1.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 23.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 3.1 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 0.8 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 3.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 3.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 9.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 3.5 | GO:0034612 | response to tumor necrosis factor(GO:0034612) |
| 0.1 | 1.6 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.8 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 3.4 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.2 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 3.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 5.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.4 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.1 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.7 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 1.0 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.0 | GO:0035696 | monocyte extravasation(GO:0035696) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 33.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 4.6 | 13.7 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 2.9 | 22.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 2.8 | 19.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 2.3 | 16.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 2.1 | 53.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 1.8 | 9.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 1.5 | 23.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 1.4 | 21.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 1.4 | 34.4 | GO:0042581 | specific granule(GO:0042581) |
| 1.2 | 23.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 1.0 | 20.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.9 | 11.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.9 | 42.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.9 | 11.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.8 | 16.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.8 | 50.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.8 | 2.4 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.8 | 6.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.7 | 6.6 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.7 | 15.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.6 | 16.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.5 | 6.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.5 | 2.8 | GO:0036019 | endolysosome(GO:0036019) |
| 0.5 | 3.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.5 | 2.3 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.5 | 3.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.5 | 1.4 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.4 | 32.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 8.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.4 | 44.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.4 | 10.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.3 | 6.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 4.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.3 | 3.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.3 | 8.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 3.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.3 | 34.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 12.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 1.8 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 10.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 21.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 3.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 14.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 36.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 2.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 11.4 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.2 | 1.5 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.2 | 4.6 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 2.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 9.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 3.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 43.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 7.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 28.0 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 8.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 13.9 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 2.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 11.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 4.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 7.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 12.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 1.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 5.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.9 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 12.1 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 13.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 2.0 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 4.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 5.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 109.3 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.5 | GO:0005903 | brush border(GO:0005903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.2 | 51.0 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 7.0 | 20.9 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 6.8 | 61.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 6.0 | 18.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 3.5 | 31.3 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 3.4 | 10.3 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 2.9 | 14.7 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 2.8 | 8.4 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 2.5 | 53.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.8 | 23.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.7 | 6.8 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 1.7 | 5.1 | GO:0002055 | adenine binding(GO:0002055) adenine phosphoribosyltransferase activity(GO:0003999) |
| 1.4 | 11.4 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) PTB domain binding(GO:0051425) |
| 1.4 | 8.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 1.4 | 8.3 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 1.2 | 5.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.2 | 3.6 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.2 | 4.6 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 1.1 | 4.6 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 1.1 | 6.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 1.0 | 36.7 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 1.0 | 37.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 1.0 | 3.0 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.9 | 6.4 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.9 | 57.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.9 | 11.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.8 | 8.8 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.7 | 43.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.6 | 3.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.6 | 9.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.6 | 16.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.6 | 7.5 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.6 | 3.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.5 | 73.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.5 | 7.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.5 | 21.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.5 | 2.8 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.4 | 3.9 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.4 | 8.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.4 | 33.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 1.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.4 | 8.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 1.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.3 | 6.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.3 | 4.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 2.0 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.3 | 8.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.3 | 20.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 15.7 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.2 | 8.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 2.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 8.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 9.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 45.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 7.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 2.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 1.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 9.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 1.7 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 14.9 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.2 | 8.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 2.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 10.2 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 1.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 24.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 10.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 3.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.5 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 14.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 22.9 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.1 | 15.9 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 3.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.9 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 6.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 3.1 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.9 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 9.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 2.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 6.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 4.2 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 5.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 27.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 13.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 4.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 1.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 2.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 10.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 7.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.7 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 2.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 8.7 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 2.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 3.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 2.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 4.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 3.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.8 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 2.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 52.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 1.1 | 61.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.8 | 58.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.7 | 67.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.6 | 23.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.6 | 57.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.5 | 40.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.5 | 11.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.4 | 19.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 15.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.3 | 17.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.3 | 12.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 2.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 10.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 1.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.2 | 14.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 8.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 8.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 8.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.2 | 5.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 14.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 10.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 22.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 2.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 4.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 34.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 1.4 | 11.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.4 | 22.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.4 | 29.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 1.1 | 56.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 1.0 | 13.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.0 | 19.2 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.9 | 16.8 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.9 | 70.6 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.7 | 25.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.7 | 57.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.7 | 16.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.6 | 8.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.5 | 8.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.5 | 2.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.4 | 32.0 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.4 | 15.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.4 | 89.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.4 | 9.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.4 | 6.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.4 | 12.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 3.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.3 | 6.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.3 | 19.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.3 | 10.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 4.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 3.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.3 | 2.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 6.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 5.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 22.0 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.2 | 5.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 9.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 2.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 1.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 3.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.6 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 13.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 8.7 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 2.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 4.2 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.1 | 2.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 2.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 6.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 4.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 4.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |