Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Taf1
Z-value: 4.14
Transcription factors associated with Taf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Taf1
|
ENSMUSG00000031314.19 | TATA-box binding protein associated factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Taf1 | mm39_v1_chrX_+_100576340_100576383 | 0.92 | 7.6e-16 | Click! |
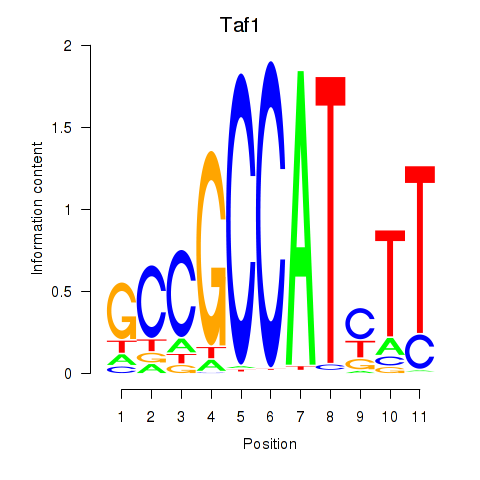
Activity profile of Taf1 motif
Sorted Z-values of Taf1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Taf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:0035037 | sperm entry(GO:0035037) |
| 3.9 | 23.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 3.5 | 10.5 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 3.5 | 13.8 | GO:0019046 | release from viral latency(GO:0019046) |
| 3.3 | 16.7 | GO:1904171 | negative regulation of bleb assembly(GO:1904171) |
| 3.2 | 9.6 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 3.0 | 9.0 | GO:1990428 | miRNA transport(GO:1990428) |
| 3.0 | 14.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 2.7 | 8.0 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 2.6 | 12.8 | GO:0090669 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 2.4 | 26.9 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 2.4 | 21.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 2.3 | 6.9 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 2.3 | 6.9 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 2.2 | 6.7 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 2.1 | 39.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 2.1 | 6.2 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 1.9 | 9.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 1.9 | 5.6 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) |
| 1.9 | 13.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 1.8 | 7.4 | GO:1904800 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 1.8 | 9.2 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 1.8 | 9.2 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 1.8 | 5.5 | GO:0016598 | protein arginylation(GO:0016598) |
| 1.8 | 8.9 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 1.8 | 8.8 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 1.7 | 5.1 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 1.7 | 6.8 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 1.6 | 4.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 1.6 | 7.9 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.6 | 10.9 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 1.6 | 7.8 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.5 | 6.1 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 1.5 | 6.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 1.5 | 7.4 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.5 | 5.9 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 1.5 | 5.9 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.5 | 4.4 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 1.5 | 5.9 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 1.4 | 7.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 1.4 | 5.6 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 1.4 | 4.2 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 1.4 | 4.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 1.4 | 5.5 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 1.3 | 10.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 1.3 | 9.4 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 1.3 | 4.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 1.3 | 2.6 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 1.3 | 22.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 1.3 | 3.9 | GO:0019085 | early viral transcription(GO:0019085) |
| 1.3 | 5.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 1.3 | 2.5 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 1.3 | 6.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 1.3 | 5.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 1.2 | 4.9 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 1.2 | 11.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 1.2 | 8.4 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 1.2 | 4.8 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 1.2 | 7.2 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 1.2 | 8.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 1.2 | 3.5 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 1.2 | 15.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.2 | 10.5 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 1.1 | 11.4 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 1.1 | 6.8 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 1.1 | 3.3 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 1.1 | 4.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.1 | 4.4 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 1.1 | 17.5 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 1.1 | 10.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 1.1 | 7.5 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 1.0 | 7.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 1.0 | 4.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 1.0 | 5.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 1.0 | 9.0 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 1.0 | 3.0 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 1.0 | 7.9 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 1.0 | 2.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 1.0 | 11.8 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 1.0 | 2.9 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 1.0 | 15.6 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 1.0 | 3.9 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 1.0 | 7.7 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.9 | 4.7 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.9 | 3.8 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.9 | 5.6 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.9 | 2.7 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.9 | 9.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.9 | 4.5 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 0.9 | 3.5 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.9 | 1.7 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.9 | 11.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.9 | 2.6 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.9 | 17.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.8 | 5.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.8 | 2.5 | GO:1902226 | negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.8 | 1.7 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.8 | 3.3 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.8 | 2.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.8 | 3.3 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.8 | 3.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.8 | 4.6 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.8 | 2.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.8 | 3.1 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.8 | 1.5 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.8 | 3.0 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.7 | 3.7 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.7 | 4.5 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.7 | 5.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.7 | 2.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.7 | 4.4 | GO:0060699 | regulation of endoribonuclease activity(GO:0060699) |
| 0.7 | 11.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.7 | 2.1 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.7 | 2.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.7 | 3.5 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.7 | 6.2 | GO:1901977 | negative regulation of cell cycle checkpoint(GO:1901977) |
| 0.7 | 2.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.7 | 12.4 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.7 | 1.4 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.7 | 1.4 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.7 | 2.7 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.7 | 4.7 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.7 | 2.0 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.7 | 3.3 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.7 | 4.6 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.7 | 4.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.6 | 5.8 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.6 | 3.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.6 | 6.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.6 | 5.8 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.6 | 4.5 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.6 | 5.7 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.6 | 23.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.6 | 2.5 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.6 | 1.9 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.6 | 2.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.6 | 1.9 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.6 | 4.4 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.6 | 1.9 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.6 | 14.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.6 | 2.5 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.6 | 6.7 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.6 | 1.8 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.6 | 4.8 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.6 | 1.8 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.6 | 3.6 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.6 | 2.4 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.6 | 4.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.6 | 5.9 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.6 | 10.5 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.6 | 2.9 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.6 | 2.9 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.6 | 2.3 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.6 | 5.7 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.6 | 1.7 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.6 | 3.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.6 | 4.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.6 | 2.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.6 | 5.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.5 | 1.6 | GO:0002588 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.5 | 2.2 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.5 | 34.8 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.5 | 3.8 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.5 | 2.7 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.5 | 3.7 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.5 | 1.6 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.5 | 4.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.5 | 3.6 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.5 | 6.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.5 | 7.7 | GO:0039529 | RIG-I signaling pathway(GO:0039529) |
| 0.5 | 11.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.5 | 2.5 | GO:0006415 | translational termination(GO:0006415) |
| 0.5 | 15.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.5 | 12.1 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.5 | 3.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.5 | 1.5 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.5 | 7.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.5 | 3.4 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.5 | 1.9 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.5 | 0.5 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.5 | 5.7 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.5 | 4.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.5 | 1.9 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.5 | 5.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.5 | 31.7 | GO:0016441 | posttranscriptional gene silencing(GO:0016441) |
| 0.5 | 22.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.4 | 3.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.4 | 4.4 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.4 | 4.4 | GO:0060969 | negative regulation of gene silencing(GO:0060969) |
| 0.4 | 7.0 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.4 | 6.1 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) |
| 0.4 | 43.0 | GO:0006413 | translational initiation(GO:0006413) |
| 0.4 | 0.9 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.4 | 3.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.4 | 7.7 | GO:1904814 | regulation of protein localization to chromosome, telomeric region(GO:1904814) |
| 0.4 | 5.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.4 | 2.5 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.4 | 2.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 | 2.8 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.4 | 2.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.4 | 1.6 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.4 | 4.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 4.6 | GO:0051461 | positive regulation of corticotropin secretion(GO:0051461) |
| 0.4 | 1.2 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.4 | 0.8 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.4 | 5.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.4 | 3.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.4 | 9.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.4 | 8.0 | GO:0036120 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.4 | 3.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.4 | 1.9 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.4 | 4.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.4 | 9.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.4 | 1.1 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.4 | 13.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.4 | 3.6 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.4 | 8.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.4 | 2.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.4 | 27.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.4 | 3.6 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.4 | 5.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.4 | 4.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.4 | 2.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.4 | 5.6 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.4 | 5.3 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.3 | 2.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.3 | 3.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 3.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.3 | 6.4 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 1.0 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.3 | 1.7 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.3 | 2.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 3.6 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.3 | 3.9 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.3 | 0.6 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.3 | 9.0 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.3 | 5.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 2.6 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.3 | 1.9 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.3 | 6.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.3 | 2.5 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.3 | 4.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.3 | 1.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 10.2 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.3 | 0.9 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.3 | 6.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.3 | 1.8 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.3 | 1.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.3 | 4.8 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.3 | 2.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.3 | 3.9 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 2.6 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.3 | 2.6 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.3 | 0.9 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 7.5 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.3 | 4.9 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.3 | 0.9 | GO:1903699 | tarsal gland development(GO:1903699) |
| 0.3 | 2.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.3 | 2.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.3 | 1.7 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.3 | 1.1 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.3 | 8.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 1.7 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.3 | 8.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.3 | 1.9 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.3 | 0.5 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.3 | 3.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 1.9 | GO:0032380 | dolichol metabolic process(GO:0019348) regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.3 | 0.3 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.3 | 1.6 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.3 | 4.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 | 1.6 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.3 | 9.3 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.3 | 1.6 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.3 | 1.0 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.3 | 2.8 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.3 | 1.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.3 | 9.0 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.3 | 7.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 1.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 3.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.3 | 7.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.3 | 6.3 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.3 | 0.5 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.3 | 1.0 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 3.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 1.5 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.2 | 3.0 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.2 | 2.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 69.5 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.2 | 1.0 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.2 | 2.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 3.5 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.2 | 3.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 0.2 | GO:0061646 | positive regulation of glutamate neurotransmitter secretion in response to membrane depolarization(GO:0061646) regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.2 | 6.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.2 | 1.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.2 | 2.7 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.2 | 1.6 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 1.8 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 7.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 7.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 0.9 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 1.3 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil activation(GO:0043307) eosinophil degranulation(GO:0043308) |
| 0.2 | 1.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.2 | 2.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 2.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 2.5 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 1.2 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 | 0.6 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 2.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 2.0 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.2 | 3.0 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.2 | 0.6 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 | 11.5 | GO:0003170 | heart valve development(GO:0003170) |
| 0.2 | 5.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 2.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 11.7 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.2 | 1.9 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.2 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 3.7 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.2 | 2.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 4.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 2.7 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.2 | 3.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 0.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 5.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.2 | 5.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 2.3 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.2 | 0.7 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.2 | 0.5 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.2 | 2.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.2 | 10.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.2 | 2.4 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.2 | 0.5 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.2 | 3.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.2 | 1.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 3.8 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.2 | 0.5 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.2 | 0.5 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 0.5 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 | 4.2 | GO:0071174 | negative regulation of mitotic metaphase/anaphase transition(GO:0045841) mitotic spindle checkpoint(GO:0071174) |
| 0.2 | 2.0 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.2 | 1.8 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 0.3 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.2 | 3.5 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.2 | 1.8 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.2 | 2.7 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.2 | 0.6 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.2 | 1.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 | 6.3 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.2 | 2.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 3.6 | GO:0007099 | centriole replication(GO:0007099) |
| 0.2 | 5.0 | GO:0038066 | p38MAPK cascade(GO:0038066) |
| 0.2 | 2.0 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.2 | 0.6 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.2 | 3.4 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.2 | 2.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 | 4.7 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.2 | 0.5 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.1 | 2.1 | GO:1903313 | positive regulation of mRNA metabolic process(GO:1903313) |
| 0.1 | 8.5 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 7.1 | GO:0017145 | stem cell division(GO:0017145) |
| 0.1 | 6.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.7 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 5.8 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 4.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.6 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 3.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 3.1 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.3 | GO:0072708 | response to sorbitol(GO:0072708) |
| 0.1 | 3.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 1.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 2.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 2.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 1.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 3.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.7 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 2.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 1.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 1.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 1.3 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 4.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 0.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 2.5 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.6 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 1.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 2.1 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.1 | 2.2 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 1.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 3.2 | GO:0007492 | endoderm development(GO:0007492) |
| 0.1 | 5.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 3.2 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 2.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.6 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.8 | GO:0019068 | virion assembly(GO:0019068) |
| 0.1 | 0.6 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 1.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 1.3 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.1 | 0.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.3 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 2.0 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 11.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 2.6 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.2 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 | 0.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 1.1 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 2.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.8 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 0.4 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 2.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 2.3 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.1 | 1.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.5 | GO:0051534 | regulation of NFAT protein import into nucleus(GO:0051532) negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 1.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 6.5 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.1 | 1.1 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.2 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.1 | 0.5 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 0.9 | GO:0010869 | regulation of receptor biosynthetic process(GO:0010869) |
| 0.1 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 2.4 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.1 | 2.8 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.1 | 0.3 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.1 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 1.5 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.1 | 1.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.5 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 0.7 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 0.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.5 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.1 | 3.6 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.6 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 1.3 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 2.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 2.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.0 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.3 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 1.7 | GO:0003281 | ventricular septum development(GO:0003281) |
| 0.0 | 2.7 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 1.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 6.6 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.3 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.2 | GO:0052205 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.1 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 3.3 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 1.2 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 2.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 2.0 | GO:0032272 | negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 1.2 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.3 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 1.5 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 1.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.3 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 | 2.9 | GO:0016071 | mRNA metabolic process(GO:0016071) |
| 0.0 | 0.8 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 1.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.5 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.0 | 0.5 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.1 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 0.5 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 1.1 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.5 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.0 | 0.7 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 2.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 2.8 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.4 | GO:0032760 | positive regulation of tumor necrosis factor production(GO:0032760) |
| 0.0 | 1.9 | GO:0007018 | microtubule-based movement(GO:0007018) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.8 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 3.0 | 20.9 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 2.8 | 22.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 2.6 | 25.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 2.3 | 6.8 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 2.0 | 7.9 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 2.0 | 13.8 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 2.0 | 5.9 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 1.9 | 3.9 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 1.9 | 7.7 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.9 | 5.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.8 | 1.8 | GO:0019034 | viral replication complex(GO:0019034) |
| 1.7 | 7.0 | GO:0001740 | Barr body(GO:0001740) |
| 1.7 | 5.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 1.6 | 11.3 | GO:0098536 | deuterosome(GO:0098536) |
| 1.6 | 11.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 1.6 | 19.2 | GO:0000796 | condensin complex(GO:0000796) |
| 1.4 | 5.7 | GO:0071920 | cleavage body(GO:0071920) |
| 1.4 | 9.9 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 1.4 | 2.8 | GO:0000974 | Prp19 complex(GO:0000974) |
| 1.3 | 6.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 1.3 | 5.3 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 1.3 | 11.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 1.3 | 16.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 1.3 | 5.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 1.3 | 2.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 1.2 | 1.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 1.2 | 4.9 | GO:0032021 | NELF complex(GO:0032021) |
| 1.2 | 22.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 1.2 | 3.6 | GO:0034455 | t-UTP complex(GO:0034455) |
| 1.2 | 3.5 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 1.2 | 7.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 1.1 | 2.2 | GO:0016589 | NURF complex(GO:0016589) |
| 1.1 | 9.6 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 1.1 | 6.4 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 1.1 | 7.4 | GO:0005638 | lamin filament(GO:0005638) |
| 1.1 | 4.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 1.0 | 15.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 1.0 | 6.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.0 | 6.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 1.0 | 9.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 1.0 | 5.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.0 | 5.7 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.9 | 7.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.9 | 1.8 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.9 | 9.7 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.9 | 7.9 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.9 | 17.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.8 | 5.9 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.8 | 31.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.8 | 2.5 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.8 | 11.4 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.8 | 8.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.8 | 5.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.8 | 4.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.8 | 4.7 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.8 | 8.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.8 | 10.0 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.8 | 6.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.8 | 11.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.7 | 3.7 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.7 | 5.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.7 | 5.0 | GO:0036396 | MIS complex(GO:0036396) |
| 0.7 | 5.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.7 | 13.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.7 | 11.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.7 | 4.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.7 | 7.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.7 | 13.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.7 | 4.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 5.1 | GO:0001652 | granular component(GO:0001652) |
| 0.6 | 2.5 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.6 | 2.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.6 | 26.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.6 | 128.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.6 | 6.0 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.6 | 8.9 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.6 | 11.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.6 | 4.6 | GO:0097422 | extrinsic component of endosome membrane(GO:0031313) tubular endosome(GO:0097422) |
| 0.6 | 46.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.6 | 13.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.6 | 2.8 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.6 | 37.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.5 | 4.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.5 | 4.7 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.5 | 1.0 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.5 | 2.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 7.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.5 | 12.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.5 | 2.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.5 | 5.9 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.5 | 5.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.5 | 6.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.5 | 2.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.5 | 37.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.5 | 2.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.5 | 7.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 5.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.5 | 3.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.4 | 4.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 2.6 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.4 | 4.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 1.6 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.4 | 4.0 | GO:0070187 | telosome(GO:0070187) |
| 0.4 | 2.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.4 | 1.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.4 | 10.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.4 | 1.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.4 | 2.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.4 | 9.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.4 | 12.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.4 | 8.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.4 | 3.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 1.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 1.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 25.9 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.3 | 3.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 5.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.3 | 2.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 1.0 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.3 | 2.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 8.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.3 | 5.2 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.3 | 10.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 3.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 24.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.3 | 2.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.3 | 12.5 | GO:0005844 | polysome(GO:0005844) |
| 0.3 | 11.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.3 | 5.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.3 | 28.6 | GO:0016605 | PML body(GO:0016605) |
| 0.3 | 4.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 7.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 1.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 1.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.3 | 6.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 0.3 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.3 | 3.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.3 | 0.8 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.3 | 1.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.3 | 4.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.2 | 1.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 8.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 1.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 33.8 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.2 | 8.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 5.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 4.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 2.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 4.8 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 1.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 1.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 2.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 8.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.2 | 2.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 1.9 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 16.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.2 | 8.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 16.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 0.5 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.2 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 0.5 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 0.5 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.2 | 2.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 5.0 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.2 | 6.5 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.2 | 3.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 1.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 4.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 95.7 | GO:0016604 | nuclear body(GO:0016604) |
| 0.2 | 1.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 19.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.2 | 0.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 1.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 6.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.7 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 3.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 4.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 1.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.3 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 2.8 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 1.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 10.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 3.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.4 | GO:0032280 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.1 | 2.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 5.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 3.2 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.1 | 2.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 5.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 2.0 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 1.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.2 | GO:0030880 | RNA polymerase complex(GO:0030880) |
| 0.1 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 7.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 2.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.7 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 37.5 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 2.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 6.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 3.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 41.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 4.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 3.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 3.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 5.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 6.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 2.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 2.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.5 | GO:0033646 | host intracellular part(GO:0033646) intracellular region of host(GO:0043656) |
| 0.0 | 5.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.1 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 23.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 2.6 | 7.9 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 2.6 | 20.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 2.4 | 9.6 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 2.4 | 9.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 2.4 | 11.9 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 2.2 | 6.5 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 1.9 | 17.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 1.9 | 13.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 1.9 | 14.8 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 1.8 | 7.2 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 1.8 | 9.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.8 | 21.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 1.7 | 20.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 1.6 | 16.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 1.6 | 1.6 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 1.6 | 4.7 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 1.5 | 10.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 1.5 | 5.9 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 1.4 | 5.5 | GO:0036033 | mediator complex binding(GO:0036033) |
| 1.3 | 8.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 1.3 | 5.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 1.3 | 5.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 1.3 | 18.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 1.2 | 7.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 1.2 | 15.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 1.1 | 5.7 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 1.1 | 7.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.0 | 4.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 1.0 | 15.6 | GO:1990405 | protein antigen binding(GO:1990405) |
| 1.0 | 9.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 1.0 | 5.7 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.9 | 9.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.9 | 5.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.9 | 6.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.9 | 60.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.9 | 2.7 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.9 | 6.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.8 | 5.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.8 | 5.9 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.8 | 5.9 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.8 | 5.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.8 | 3.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.7 | 3.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.7 | 5.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.7 | 48.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.7 | 10.9 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.7 | 13.8 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.7 | 5.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.7 | 10.4 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.7 | 10.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.7 | 4.7 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.7 | 10.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.7 | 5.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.6 | 6.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.6 | 3.8 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.6 | 4.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.6 | 5.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.6 | 6.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.6 | 3.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.6 | 5.7 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.6 | 4.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.6 | 11.8 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.6 | 1.7 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.5 | 1.0 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.5 | 18.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.5 | 1.6 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.5 | 4.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.5 | 1.6 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.5 | 4.6 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.5 | 2.1 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.5 | 4.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.5 | 3.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.5 | 1.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.5 | 11.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.5 | 1.5 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.5 | 2.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.5 | 1.5 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.5 | 2.5 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.5 | 3.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.5 | 1.0 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.5 | 2.4 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.5 | 1.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.5 | 1.9 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) |
| 0.5 | 14.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.5 | 1.9 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.5 | 2.8 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.5 | 4.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.5 | 3.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.5 | 2.7 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.5 | 3.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.4 | 4.4 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.4 | 8.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.4 | 5.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.4 | 2.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 4.6 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.4 | 2.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 4.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.4 | 1.2 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.4 | 3.5 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.4 | 18.7 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.4 | 3.5 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.4 | 3.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.4 | 23.8 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.4 | 1.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.4 | 1.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 1.8 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.4 | 2.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.4 | 3.9 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.3 | 1.0 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.3 | 1.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 1.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.3 | 8.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 0.3 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.3 | 1.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 0.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.3 | 3.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 3.5 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.3 | 6.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 7.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 1.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.3 | 32.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.3 | 5.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 3.6 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 1.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 8.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.3 | 3.0 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.3 | 2.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.3 | 3.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.3 | 44.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.3 | 2.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.3 | 5.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 12.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 0.8 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.3 | 2.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 1.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 1.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.3 | 16.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 7.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 6.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 0.8 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 4.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.3 | 11.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.3 | 3.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 2.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 10.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.3 | 5.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 6.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 3.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 5.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 1.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.2 | 13.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 1.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 6.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 21.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 1.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 4.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 2.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 9.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.2 | 9.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 4.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 5.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 5.8 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.2 | 6.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 4.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 0.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 4.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 46.6 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.2 | 5.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.2 | 0.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 4.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 8.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 71.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.2 | 10.7 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.2 | 6.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 6.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 7.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 4.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 2.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 0.5 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.2 | 2.6 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 3.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 0.5 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.2 | 1.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 4.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 3.6 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 2.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 3.2 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.2 | 2.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.2 | 6.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 0.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 13.2 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.2 | 0.5 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.1 | 10.6 | GO:0034212 | peptide N-acetyltransferase activity(GO:0034212) |
| 0.1 | 0.4 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 6.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 10.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 7.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 198.0 | GO:0003723 | RNA binding(GO:0003723) |
| 0.1 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 2.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.4 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 9.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 2.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 2.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 3.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 3.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 10.7 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 1.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 2.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.6 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.5 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.5 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 5.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 2.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 4.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 5.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.7 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 2.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 6.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 2.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.6 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 8.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 2.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.5 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 3.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 2.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 3.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 2.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 3.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 1.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.8 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 7.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0008310 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 7.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 4.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 20.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 21.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 4.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 1.4 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 3.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 9.7 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.4 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 2.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.8 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 7.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 2.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.5 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.4 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 3.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.6 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 2.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 5.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 1.0 | 59.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.6 | 38.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.6 | 10.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.6 | 5.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.5 | 33.8 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.5 | 16.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.4 | 8.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.4 | 17.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.4 | 12.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.3 | 34.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.3 | 13.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.3 | 3.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.3 | 15.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 6.8 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 16.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 8.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 24.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 9.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 9.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 12.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 13.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 9.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 9.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.2 | 2.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 8.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 6.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 4.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 2.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 8.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 3.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 12.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 7.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 5.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 5.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 4.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 7.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 9.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 4.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 5.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 1.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 4.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 18.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 4.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.8 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 5.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 1.5 | 15.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 1.2 | 62.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 1.1 | 35.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.8 | 7.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.8 | 12.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.7 | 26.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.7 | 9.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.7 | 9.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.7 | 75.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.7 | 14.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.6 | 14.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.6 | 23.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.6 | 13.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.6 | 4.0 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.6 | 4.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.5 | 12.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.5 | 3.0 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.5 | 3.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.5 | 40.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.5 | 11.8 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.5 | 11.7 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.5 | 5.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.5 | 9.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.5 | 6.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.5 | 55.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.4 | 16.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.4 | 27.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.4 | 6.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 3.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.4 | 32.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.4 | 24.6 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.4 | 8.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.4 | 8.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 6.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 3.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 10.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 3.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.3 | 14.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.3 | 4.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 6.1 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.3 | 4.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 12.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.3 | 10.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.3 | 1.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.3 | 15.5 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.3 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.3 | 8.0 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.3 | 8.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.2 | 4.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 4.9 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 4.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 1.7 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.2 | 3.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 8.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 0.7 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 5.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 4.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 2.5 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.2 | 4.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 5.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 3.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 5.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 2.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.2 | 3.9 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.2 | 5.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 5.9 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.1 | 3.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 4.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 15.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.9 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.1 | 2.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 4.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 2.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 4.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 3.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 7.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.8 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 9.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 9.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 23.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 6.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.8 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 3.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 1.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |