Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
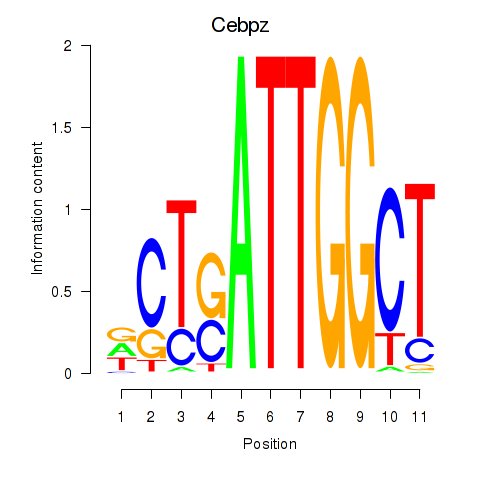
Results for Ybx1_Nfya_Nfyb_Nfyc_Cebpz
Z-value: 6.00
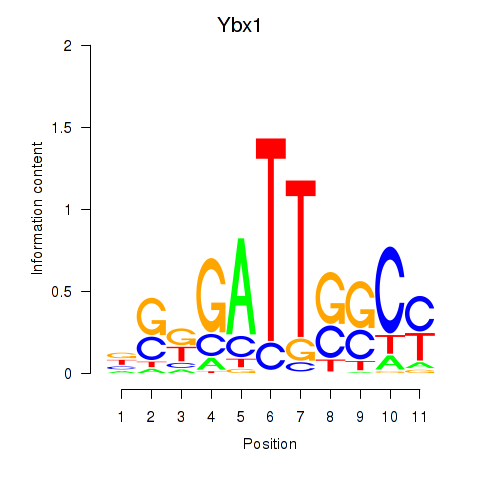
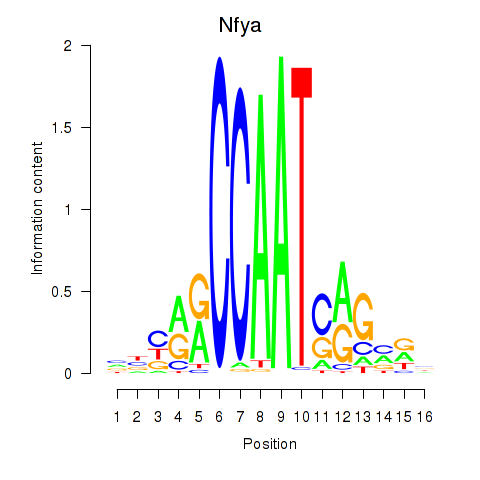
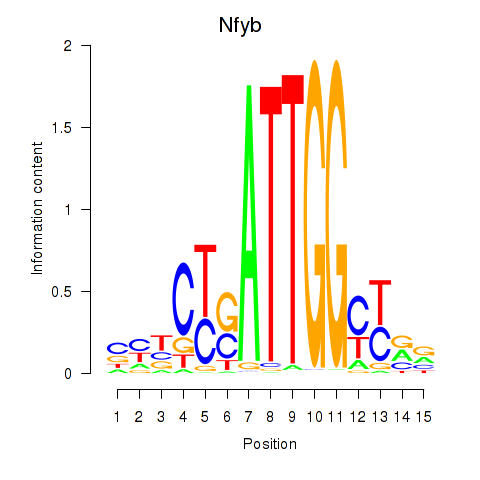
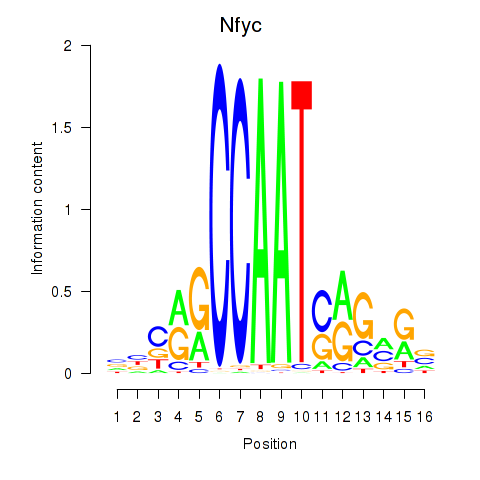
Transcription factors associated with Ybx1_Nfya_Nfyb_Nfyc_Cebpz
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ybx1
|
ENSMUSG00000028639.15 | Y box protein 1 |
|
Nfya
|
ENSMUSG00000023994.14 | nuclear transcription factor-Y alpha |
|
Nfyb
|
ENSMUSG00000020248.19 | nuclear transcription factor-Y beta |
|
Nfyc
|
ENSMUSG00000032897.18 | nuclear transcription factor-Y gamma |
|
Cebpz
|
ENSMUSG00000024081.10 | CCAAT/enhancer binding protein zeta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfyc | mm39_v1_chr4_-_120672900_120672959 | 0.84 | 9.6e-11 | Click! |
| Nfya | mm39_v1_chr17_-_48716756_48716934 | 0.81 | 3.0e-09 | Click! |
| Ybx1 | mm39_v1_chr4_-_119151717_119151801 | 0.75 | 1.4e-07 | Click! |
| Nfyb | mm39_v1_chr10_-_82599967_82599991 | 0.55 | 4.9e-04 | Click! |
| Cebpz | mm39_v1_chr17_-_79244460_79244504 | 0.39 | 2.0e-02 | Click! |
Activity profile of Ybx1_Nfya_Nfyb_Nfyc_Cebpz motif
Sorted Z-values of Ybx1_Nfya_Nfyb_Nfyc_Cebpz motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Ybx1_Nfya_Nfyb_Nfyc_Cebpz
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.2 | 61.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 12.2 | 36.5 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 9.8 | 29.3 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 9.8 | 29.3 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 9.7 | 29.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 9.5 | 38.2 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 9.3 | 93.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 9.3 | 93.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 8.5 | 93.0 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 8.4 | 25.2 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 8.2 | 16.5 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 7.8 | 62.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 7.6 | 22.7 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 7.5 | 22.5 | GO:0015825 | L-serine transport(GO:0015825) |
| 7.0 | 42.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 7.0 | 56.1 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 6.9 | 34.6 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 6.7 | 100.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 6.5 | 19.5 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 6.3 | 25.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 6.1 | 18.4 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 6.1 | 18.2 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 6.0 | 18.0 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 5.4 | 21.5 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 5.4 | 16.1 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 5.3 | 15.9 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 5.3 | 15.8 | GO:0070194 | synaptonemal complex disassembly(GO:0070194) |
| 5.2 | 20.6 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 4.9 | 58.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 4.9 | 19.4 | GO:0006272 | leading strand elongation(GO:0006272) |
| 4.7 | 18.8 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 4.5 | 18.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 4.4 | 26.3 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 4.3 | 12.9 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 4.2 | 21.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 4.2 | 33.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 4.2 | 12.6 | GO:0034117 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 4.1 | 78.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 4.0 | 12.0 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 4.0 | 11.9 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 4.0 | 23.8 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 3.9 | 15.7 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 3.8 | 15.4 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 3.8 | 37.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 3.7 | 22.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 3.7 | 73.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 3.6 | 54.6 | GO:0048102 | autophagic cell death(GO:0048102) |
| 3.6 | 10.9 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 3.6 | 43.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 3.6 | 21.5 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 3.5 | 17.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 3.4 | 20.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 3.3 | 19.9 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 3.3 | 10.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 3.3 | 19.8 | GO:1903182 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 3.3 | 19.6 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 3.2 | 12.9 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 3.2 | 16.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 3.2 | 6.4 | GO:0072708 | response to sorbitol(GO:0072708) |
| 3.2 | 9.5 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 3.2 | 41.3 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 3.2 | 28.5 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 3.1 | 18.6 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 3.1 | 18.5 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 3.1 | 27.7 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 3.0 | 24.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 3.0 | 27.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 3.0 | 33.1 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 3.0 | 29.8 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 3.0 | 17.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 2.9 | 23.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 2.9 | 5.8 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 2.9 | 5.8 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 2.9 | 71.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 2.8 | 42.6 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 2.8 | 126.5 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 2.7 | 8.0 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 2.6 | 13.2 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 2.5 | 12.4 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 2.5 | 2.5 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 2.5 | 7.4 | GO:0070376 | regulation of ERK5 cascade(GO:0070376) |
| 2.4 | 7.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 2.3 | 18.0 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 2.3 | 36.0 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 2.2 | 15.7 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 2.2 | 17.8 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 2.2 | 6.7 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 2.2 | 2.2 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 2.1 | 30.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 2.1 | 10.6 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 2.1 | 6.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 2.1 | 8.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 2.1 | 10.4 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 2.1 | 10.3 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 2.0 | 12.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 2.0 | 8.1 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 2.0 | 16.1 | GO:0044838 | cell quiescence(GO:0044838) |
| 2.0 | 4.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 2.0 | 28.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 2.0 | 21.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 2.0 | 5.9 | GO:1990751 | regulation of Schwann cell chemotaxis(GO:1904266) positive regulation of Schwann cell chemotaxis(GO:1904268) Schwann cell chemotaxis(GO:1990751) |
| 2.0 | 23.5 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 1.9 | 13.6 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 1.9 | 9.7 | GO:0051595 | response to methylglyoxal(GO:0051595) |
| 1.9 | 7.7 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.9 | 28.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 1.9 | 34.2 | GO:0043486 | histone exchange(GO:0043486) |
| 1.9 | 20.8 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 1.9 | 3.7 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 1.8 | 5.5 | GO:0097037 | heme export(GO:0097037) |
| 1.8 | 16.4 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 1.8 | 14.4 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.8 | 9.0 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 1.8 | 5.4 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 1.8 | 10.7 | GO:0007144 | female meiosis I(GO:0007144) |
| 1.8 | 19.6 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 1.8 | 12.3 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 1.7 | 12.2 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 1.7 | 5.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 1.7 | 5.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 1.6 | 1.6 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 1.6 | 25.7 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 1.6 | 16.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 1.5 | 26.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 1.5 | 4.6 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 1.5 | 9.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.5 | 7.6 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 1.5 | 5.9 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 1.5 | 30.6 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 1.5 | 10.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 1.4 | 4.3 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 1.4 | 21.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 1.4 | 11.5 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 1.4 | 2.8 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.4 | 4.2 | GO:0070203 | regulation of establishment of protein localization to chromosome(GO:0070202) regulation of establishment of protein localization to telomere(GO:0070203) |
| 1.4 | 5.6 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 1.4 | 18.0 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 1.4 | 9.7 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 1.4 | 30.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 1.4 | 24.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 1.3 | 14.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 1.3 | 14.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.3 | 6.6 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 1.3 | 36.7 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 1.3 | 11.5 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 1.3 | 5.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 1.3 | 5.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 1.2 | 3.7 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 1.2 | 68.0 | GO:0048821 | erythrocyte development(GO:0048821) |
| 1.2 | 11.0 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 1.2 | 7.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 1.2 | 3.6 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.2 | 8.5 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 1.2 | 10.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 1.2 | 6.0 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 1.2 | 3.6 | GO:0016131 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 1.2 | 4.7 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 1.2 | 19.8 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 1.1 | 61.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 1.1 | 3.4 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 1.1 | 4.5 | GO:0010286 | heat acclimation(GO:0010286) |
| 1.1 | 25.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 1.1 | 2.2 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 1.1 | 6.7 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 1.1 | 4.5 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 1.1 | 4.4 | GO:2001271 | oxygen metabolic process(GO:0072592) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 1.1 | 5.5 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.1 | 5.5 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.1 | 8.7 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.1 | 9.8 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 1.1 | 1.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 1.0 | 10.4 | GO:0021995 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 1.0 | 10.3 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.0 | 19.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 1.0 | 4.9 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.0 | 3.9 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 1.0 | 13.7 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 1.0 | 7.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 1.0 | 9.5 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.9 | 9.4 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.9 | 2.8 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.9 | 4.7 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.9 | 2.8 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.9 | 4.6 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.9 | 2.7 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.9 | 3.6 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.9 | 18.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.9 | 12.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.9 | 13.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.9 | 0.9 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.9 | 21.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.9 | 14.9 | GO:1904872 | regulation of telomerase RNA localization to Cajal body(GO:1904872) |
| 0.9 | 4.3 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.9 | 7.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.9 | 3.5 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.9 | 4.3 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.9 | 22.4 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.9 | 5.2 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.8 | 12.7 | GO:0007099 | centriole replication(GO:0007099) |
| 0.8 | 6.7 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.8 | 3.3 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.8 | 18.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.8 | 2.4 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.8 | 8.1 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.8 | 5.6 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.8 | 4.8 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.8 | 7.1 | GO:2000774 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.8 | 11.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.8 | 2.4 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.8 | 2.4 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.8 | 2.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.8 | 6.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.8 | 3.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.8 | 5.4 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.8 | 4.6 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.8 | 3.8 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.8 | 49.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.8 | 0.8 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.8 | 3.0 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.8 | 6.8 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.7 | 5.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.7 | 5.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.7 | 6.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.7 | 5.1 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.7 | 3.6 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.7 | 26.5 | GO:0031050 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.7 | 1.4 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.7 | 25.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.7 | 2.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.7 | 34.5 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.7 | 8.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.7 | 4.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.7 | 2.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.7 | 1.4 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.7 | 0.7 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.7 | 14.6 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.6 | 6.5 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.6 | 5.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.6 | 6.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.6 | 19.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.6 | 1.9 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.6 | 3.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.6 | 5.6 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.6 | 73.3 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.6 | 3.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.6 | 3.7 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.6 | 2.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.6 | 3.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.6 | 5.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.6 | 3.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.6 | 1.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.6 | 10.3 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.6 | 29.1 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.6 | 19.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.6 | 113.8 | GO:0044843 | cell cycle G1/S phase transition(GO:0044843) |
| 0.6 | 24.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.6 | 9.5 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.6 | 1.8 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.6 | 4.1 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.6 | 4.6 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.6 | 1.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.6 | 6.9 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.6 | 2.8 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.6 | 9.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.6 | 3.9 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 0.6 | 5.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.5 | 32.6 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.5 | 3.3 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.5 | 5.4 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.5 | 4.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.5 | 6.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.5 | 7.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.5 | 2.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.5 | 6.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.5 | 56.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.5 | 2.6 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.5 | 4.6 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.5 | 15.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.5 | 4.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.5 | 2.5 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.5 | 3.6 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.5 | 5.6 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.5 | 1.0 | GO:0036034 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.5 | 1.0 | GO:2000612 | regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.5 | 0.5 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.5 | 2.0 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.5 | 9.3 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.5 | 1.9 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.5 | 2.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.5 | 2.4 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.5 | 12.1 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.5 | 2.4 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.5 | 1.9 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.5 | 6.1 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.5 | 7.5 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.5 | 2.8 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.5 | 2.3 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.5 | 5.1 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.5 | 2.3 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.4 | 3.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.4 | 7.5 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.4 | 3.5 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.4 | 12.2 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.4 | 15.2 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.4 | 3.8 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.4 | 5.0 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.4 | 2.1 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.4 | 1.7 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.4 | 0.4 | GO:0048866 | stem cell fate specification(GO:0048866) |
| 0.4 | 6.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.4 | 3.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.4 | 4.8 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.4 | 9.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.4 | 1.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.4 | 3.2 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.4 | 6.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.4 | 3.8 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.4 | 1.5 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.4 | 4.9 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.4 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.4 | 4.4 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.4 | 1.8 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.4 | 5.8 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.4 | 9.0 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.4 | 0.7 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.4 | 7.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.4 | 1.4 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.4 | 2.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.3 | 1.7 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.3 | 26.7 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.3 | 2.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 1.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.3 | 1.3 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) positive regulation of progesterone secretion(GO:2000872) |
| 0.3 | 1.3 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.3 | 3.0 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.3 | 1.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.3 | 1.0 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.3 | 4.5 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.3 | 1.0 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.3 | 10.9 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.3 | 1.0 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.3 | 4.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.3 | 1.9 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.3 | 2.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.3 | 8.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.3 | 3.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.3 | 11.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.3 | 1.8 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.3 | 1.5 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.3 | 4.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.3 | 2.0 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.3 | 2.3 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.3 | 0.9 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.3 | 4.3 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.3 | 0.6 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.3 | 0.8 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.3 | 9.8 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.3 | 2.5 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.3 | 4.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.3 | 6.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.3 | 4.6 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.3 | 10.4 | GO:0009620 | response to fungus(GO:0009620) |
| 0.3 | 4.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.3 | 2.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.3 | 2.6 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.3 | 2.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.3 | 0.5 | GO:2000836 | positive regulation of androgen secretion(GO:2000836) positive regulation of testosterone secretion(GO:2000845) |
| 0.3 | 1.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 2.1 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.3 | 0.3 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.3 | 1.0 | GO:1902080 | regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.2 | 1.7 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.2 | 1.9 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 1.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.2 | 1.9 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.2 | 10.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 1.9 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 0.9 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.2 | 6.4 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.2 | 1.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.5 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) |
| 0.2 | 2.0 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 | 4.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 2.6 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 0.6 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.2 | 10.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 0.6 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.2 | 0.4 | GO:0070099 | regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.2 | 4.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 79.6 | GO:0051301 | cell division(GO:0051301) |
| 0.2 | 2.3 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.2 | 3.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 1.0 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 2.7 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.2 | 21.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.2 | 1.4 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.2 | 0.8 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 0.2 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.2 | 2.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 3.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 1.6 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 2.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 0.8 | GO:2000983 | regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.2 | 1.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 2.6 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.2 | 3.9 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.2 | 2.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.2 | 1.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 1.0 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.2 | 1.5 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.2 | 0.7 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 0.7 | GO:0051031 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 3.8 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.2 | 0.3 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.2 | 0.8 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 3.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.2 | 8.2 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.2 | 0.6 | GO:1902047 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 1.7 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 | 0.5 | GO:1902473 | regulation of protein localization to synapse(GO:1902473) |
| 0.2 | 1.7 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.2 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 0.8 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 1.2 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.2 | 4.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.2 | 0.5 | GO:0030200 | proteoglycan catabolic process(GO:0030167) heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.9 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.4 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 3.9 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 1.0 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 3.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 6.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.5 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 | 0.8 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 1.5 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 3.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 2.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 6.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 2.6 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.1 | 9.8 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.1 | 4.6 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 1.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 2.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 1.7 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 | 2.2 | GO:0055098 | response to lipoprotein particle(GO:0055094) response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.9 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 0.9 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.1 | 1.4 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.1 | 0.9 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.5 | GO:0071426 | ribonucleoprotein complex export from nucleus(GO:0071426) |
| 0.1 | 0.3 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 1.7 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 8.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 0.8 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.1 | 4.5 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 2.0 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 4.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.3 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.3 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 6.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 1.4 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 | 1.3 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 10.7 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.1 | 2.0 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 2.6 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.0 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.3 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 0.2 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 1.4 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.1 | 0.2 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.1 | 1.9 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 0.6 | GO:0070587 | negative regulation of heterotypic cell-cell adhesion(GO:0034115) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 9.2 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 0.2 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.1 | 0.1 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.1 | 2.6 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 0.6 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 0.3 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.2 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 3.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.3 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 0.2 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 2.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 1.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 1.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.4 | GO:0031274 | regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.4 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 1.0 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.1 | GO:0003183 | mitral valve development(GO:0003174) mitral valve morphogenesis(GO:0003183) |
| 0.0 | 0.9 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0030719 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.0 | 1.4 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.3 | GO:0030913 | protein localization to paranode region of axon(GO:0002175) paranodal junction assembly(GO:0030913) |
| 0.0 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 1.3 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 1.0 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.0 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.4 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.5 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.9 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.8 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.0 | 1.0 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) |
| 0.0 | 0.4 | GO:0090314 | positive regulation of protein targeting to membrane(GO:0090314) |
| 0.0 | 0.3 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 2.0 | GO:0031623 | receptor internalization(GO:0031623) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.3 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.9 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.4 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:0090673 | endothelial cell-matrix adhesion(GO:0090673) |
| 0.0 | 0.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.7 | 93.4 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 15.1 | 90.6 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 14.8 | 74.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 13.8 | 82.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 12.1 | 48.3 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 9.7 | 29.2 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 9.6 | 28.9 | GO:1990031 | pinceau fiber(GO:1990031) |
| 9.4 | 18.8 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 8.7 | 86.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 7.2 | 57.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 6.3 | 25.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 6.1 | 24.6 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 6.0 | 24.2 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 5.9 | 29.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 5.4 | 86.5 | GO:0042555 | MCM complex(GO:0042555) |
| 5.0 | 15.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 5.0 | 15.0 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 5.0 | 65.0 | GO:0005818 | aster(GO:0005818) |
| 4.8 | 14.5 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 4.8 | 42.9 | GO:0045298 | tubulin complex(GO:0045298) |
| 4.5 | 17.9 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 4.1 | 45.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 3.9 | 3.9 | GO:0061574 | ASAP complex(GO:0061574) |
| 3.8 | 37.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 3.8 | 18.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 3.7 | 116.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 3.5 | 3.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 3.4 | 47.3 | GO:0001939 | female pronucleus(GO:0001939) |
| 3.3 | 26.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 3.3 | 19.8 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 3.3 | 125.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 3.1 | 36.9 | GO:0034709 | methylosome(GO:0034709) |
| 2.8 | 8.3 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 2.7 | 32.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 2.6 | 15.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 2.5 | 25.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 2.4 | 9.8 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 2.2 | 20.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 2.1 | 32.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 2.1 | 6.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 2.0 | 8.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 2.0 | 17.8 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 1.9 | 13.6 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 1.9 | 38.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 1.9 | 13.6 | GO:0005638 | lamin filament(GO:0005638) |
| 1.9 | 5.7 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 1.9 | 28.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 1.9 | 11.1 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 1.8 | 5.5 | GO:0071547 | piP-body(GO:0071547) |
| 1.8 | 23.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 1.8 | 74.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 1.7 | 22.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 1.6 | 31.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 1.6 | 4.8 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 1.6 | 17.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 1.6 | 10.9 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 1.5 | 4.5 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 1.5 | 12.0 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 1.5 | 5.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.5 | 94.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 1.4 | 5.8 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 1.4 | 14.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.4 | 15.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 1.4 | 12.5 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 1.4 | 9.6 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 1.4 | 13.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 1.4 | 166.1 | GO:0000776 | kinetochore(GO:0000776) |
| 1.3 | 18.6 | GO:0045120 | pronucleus(GO:0045120) |
| 1.3 | 59.2 | GO:0005657 | replication fork(GO:0005657) |
| 1.3 | 5.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 1.2 | 154.0 | GO:0005814 | centriole(GO:0005814) |
| 1.2 | 3.6 | GO:0008278 | cohesin complex(GO:0008278) |
| 1.2 | 3.6 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 1.2 | 6.0 | GO:0031251 | PAN complex(GO:0031251) |
| 1.2 | 5.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 1.1 | 4.6 | GO:0005687 | U4 snRNP(GO:0005687) |
| 1.1 | 60.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 1.1 | 5.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.1 | 3.3 | GO:0033503 | HULC complex(GO:0033503) |
| 1.1 | 11.6 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 1.0 | 4.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.0 | 15.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 1.0 | 14.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 1.0 | 17.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 1.0 | 5.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.0 | 3.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 1.0 | 30.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 1.0 | 2.0 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.9 | 1.9 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.9 | 2.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.9 | 10.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.9 | 11.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.9 | 21.6 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.9 | 24.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.9 | 0.9 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.9 | 12.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.9 | 4.3 | GO:0070187 | telosome(GO:0070187) |
| 0.9 | 13.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.9 | 2.6 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.9 | 12.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.9 | 21.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.9 | 28.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.9 | 17.0 | GO:0070938 | contractile ring(GO:0070938) |
| 0.8 | 6.7 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.8 | 18.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.8 | 11.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.8 | 3.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.8 | 2.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.7 | 4.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.7 | 126.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.7 | 2.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.7 | 2.1 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.7 | 0.7 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.7 | 2.8 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.7 | 3.5 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.7 | 4.9 | GO:0036396 | MIS complex(GO:0036396) |
| 0.7 | 15.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.7 | 10.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.7 | 3.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.7 | 7.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.7 | 44.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.6 | 6.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.6 | 25.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.6 | 35.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.6 | 10.7 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.6 | 10.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.6 | 55.1 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.6 | 3.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.6 | 12.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.6 | 5.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.6 | 52.5 | GO:0016605 | PML body(GO:0016605) |
| 0.6 | 26.4 | GO:0005819 | spindle(GO:0005819) |
| 0.6 | 3.4 | GO:0034448 | EGO complex(GO:0034448) |
| 0.6 | 226.4 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.5 | 8.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.5 | 31.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.5 | 6.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.5 | 2.9 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.5 | 36.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.5 | 2.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.5 | 2.9 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.5 | 2.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.5 | 11.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.5 | 1.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.5 | 1.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.5 | 11.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.5 | 7.2 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.5 | 12.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.4 | 0.4 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.4 | 3.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.4 | 2.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.4 | 13.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.4 | 19.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.4 | 37.7 | GO:0030496 | midbody(GO:0030496) |
| 0.4 | 18.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.4 | 8.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.4 | 3.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.4 | 3.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.4 | 1.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.4 | 39.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.4 | 2.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.4 | 4.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.4 | 4.0 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.4 | 9.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 21.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.4 | 2.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.3 | 3.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 2.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 2.7 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.3 | 0.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 11.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.3 | 2.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 20.4 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.3 | 40.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.3 | 1.7 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.3 | 3.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 2.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.3 | 1.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 2.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 1.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 1.7 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 2.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 0.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 1.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.2 | 6.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 0.7 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.2 | 6.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 3.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 1.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 1.8 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.2 | 2.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 14.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 13.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 1.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 6.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 51.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.2 | 2.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 1.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 53.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.2 | 8.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 1.0 | GO:0097443 | sorting endosome(GO:0097443) cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 0.5 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.2 | 1.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 3.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 2.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 3.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.9 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.6 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 1.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 3.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 5.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.8 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 17.5 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 4.7 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 2.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 0.6 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 4.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.9 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 8.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 7.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 14.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.6 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 1.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.4 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 7.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 3.5 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.1 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 4.0 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 24.7 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 0.9 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 7.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 4.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 38.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 2.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 4.5 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.6 | 74.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 15.9 | 63.5 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 11.0 | 65.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 8.8 | 88.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 7.4 | 59.5 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 6.7 | 33.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 6.5 | 39.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 6.1 | 18.3 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 5.8 | 23.3 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 5.5 | 27.3 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 5.4 | 16.1 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 4.8 | 19.4 | GO:0030519 | snoRNP binding(GO:0030519) |
| 4.7 | 14.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 4.7 | 37.8 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 4.7 | 23.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 4.7 | 14.1 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 4.6 | 13.9 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 4.4 | 26.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 4.4 | 88.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 4.3 | 21.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 4.3 | 21.3 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 4.2 | 76.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 4.1 | 37.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 4.0 | 12.0 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 3.9 | 27.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 3.3 | 13.3 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 3.3 | 19.8 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 3.2 | 9.5 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 3.2 | 15.9 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 3.2 | 12.7 | GO:0048030 | disaccharide binding(GO:0048030) |
| 3.1 | 3.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 2.9 | 20.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 2.9 | 49.0 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 2.8 | 16.7 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 2.8 | 2.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 2.7 | 10.8 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 2.6 | 34.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 2.6 | 18.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 2.6 | 84.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 2.6 | 17.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 2.5 | 7.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 2.4 | 22.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 2.3 | 30.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 2.3 | 24.8 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 2.1 | 6.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 2.1 | 8.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 2.1 | 20.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 2.1 | 6.2 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 2.0 | 6.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 2.0 | 8.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 1.9 | 7.5 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 1.8 | 5.5 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 1.8 | 12.7 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.8 | 5.3 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 1.7 | 5.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 1.7 | 6.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.7 | 5.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 1.7 | 22.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 1.7 | 5.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 1.7 | 69.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 1.7 | 23.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 1.7 | 3.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 1.7 | 6.6 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.6 | 11.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 1.6 | 58.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 1.6 | 4.7 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 1.5 | 7.7 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.5 | 13.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 1.5 | 170.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 1.5 | 9.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.5 | 5.8 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 1.4 | 5.7 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 1.4 | 4.3 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 1.4 | 4.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 1.4 | 5.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 1.4 | 5.5 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 1.4 | 4.1 | GO:0008456 | alpha-N-acetylgalactosaminidase activity(GO:0008456) |
| 1.3 | 21.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 1.3 | 5.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.3 | 14.4 | GO:0043142 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 1.3 | 69.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 1.3 | 5.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 1.3 | 6.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.3 | 5.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 1.2 | 3.7 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.2 | 18.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.2 | 2.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 1.2 | 6.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 1.2 | 6.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 1.2 | 32.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 1.2 | 3.6 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 1.2 | 7.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 1.1 | 5.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.1 | 4.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 1.1 | 4.5 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 1.1 | 3.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 1.1 | 6.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 1.1 | 19.4 | GO:0008430 | selenium binding(GO:0008430) |
| 1.1 | 4.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 1.1 | 48.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 1.1 | 236.7 | GO:0042393 | histone binding(GO:0042393) |
| 1.1 | 6.4 | GO:0070728 | leucine binding(GO:0070728) |
| 1.0 | 3.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.0 | 3.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 1.0 | 4.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 1.0 | 3.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.0 | 4.0 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 1.0 | 4.8 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 1.0 | 2.9 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.9 | 3.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.9 | 37.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.9 | 2.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.9 | 2.8 | GO:0004512 | inositol-3-phosphate synthase activity(GO:0004512) intramolecular lyase activity(GO:0016872) |
| 0.9 | 7.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.9 | 2.8 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.9 | 2.8 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.9 | 17.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.9 | 2.7 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.9 | 25.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.9 | 4.5 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.9 | 4.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.9 | 12.0 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.9 | 47.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.9 | 7.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.8 | 6.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.8 | 2.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.8 | 6.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.8 | 31.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.8 | 4.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.8 | 6.5 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.8 | 5.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.8 | 4.8 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.8 | 3.2 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.8 | 10.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.8 | 5.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.8 | 9.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.8 | 2.3 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.8 | 1.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.7 | 10.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.7 | 2.9 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.7 | 16.0 | GO:0010181 | FMN binding(GO:0010181) |
| 0.7 | 2.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.7 | 12.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.7 | 12.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.7 | 5.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.7 | 21.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.7 | 2.7 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.7 | 10.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.7 | 3.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.6 | 9.1 | GO:0070990 | snRNP binding(GO:0070990) |
| 0.6 | 5.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.6 | 12.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.6 | 7.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.6 | 3.7 | GO:0089720 | caspase binding(GO:0089720) |
| 0.6 | 9.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.6 | 1.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.6 | 4.8 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.6 | 2.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.6 | 2.9 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.6 | 14.9 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.6 | 20.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.6 | 4.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.6 | 6.7 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.6 | 3.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.6 | 13.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 4.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.5 | 26.2 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.5 | 2.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.5 | 4.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.5 | 3.7 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.5 | 11.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.5 | 12.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.5 | 21.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.5 | 2.6 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.5 | 11.9 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.5 | 2.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.5 | 76.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.5 | 1.0 | GO:0032357 | oxidized purine DNA binding(GO:0032357) |
| 0.5 | 2.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.5 | 6.8 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.5 | 2.9 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.5 | 4.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 4.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.5 | 1.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.5 | 43.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.5 | 4.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.5 | 35.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.5 | 2.8 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.5 | 10.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.5 | 3.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.4 | 2.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.4 | 3.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.4 | 2.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 4.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.4 | 3.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.4 | 2.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.4 | 2.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 1.6 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.4 | 7.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.4 | 3.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.4 | 9.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.4 | 6.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 1.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.4 | 88.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.4 | 1.1 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.4 | 1.5 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.4 | 20.0 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.3 | 2.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 29.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.3 | 19.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 5.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 4.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 12.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.3 | 3.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.3 | 1.6 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.3 | 3.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 9.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.3 | 8.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 8.5 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.3 | 12.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.3 | 6.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 10.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 1.5 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.3 | 1.8 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.3 | 0.9 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.3 | 67.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.3 | 0.6 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.3 | 1.9 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.3 | 6.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.3 | 1.7 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 1.6 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.3 | 7.2 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.3 | 1.0 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.2 | 2.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 1.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 1.0 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.2 | 129.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.2 | 4.6 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 12.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 1.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 1.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 0.9 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 8.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 10.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 0.6 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.2 | 7.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 1.4 | GO:0030899 | troponin C binding(GO:0030172) calcium-dependent ATPase activity(GO:0030899) troponin I binding(GO:0031013) |
| 0.2 | 1.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 5.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 8.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 0.8 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 0.8 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.2 | 5.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 3.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.2 | 5.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 31.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 1.6 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 3.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 8.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 64.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 0.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 31.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 2.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 5.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 15.9 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.2 | 2.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 0.8 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.8 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 1.6 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.2 | 0.6 | GO:0042979 | ornithine decarboxylase inhibitor activity(GO:0008073) ornithine decarboxylase regulator activity(GO:0042979) |
| 0.2 | 0.9 | GO:0052723 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 1.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.9 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 10.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 1.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 1.5 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 12.0 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 2.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 1.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.4 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.2 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 7.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 5.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 16.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 1.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.8 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 1.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.1 | GO:0003681 | bent DNA binding(GO:0003681) |
| 0.1 | 0.3 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 0.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 4.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 2.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 1.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 30.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 12.2 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 0.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.0 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 0.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 5.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 29.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 2.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 1.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 1.7 | GO:0016896 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.4 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 2.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.1 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.0 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 2.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.0 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 1.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.7 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0031811 | beta-1 adrenergic receptor binding(GO:0031697) G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 348.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 4.4 | 82.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 4.3 | 199.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 3.0 | 171.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 2.3 | 100.0 | PID ATR PATHWAY | ATR signaling pathway |
| 2.1 | 41.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 1.6 | 53.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 1.4 | 115.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 1.4 | 13.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 1.4 | 41.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 1.3 | 33.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 1.2 | 31.3 | PID MYC PATHWAY | C-MYC pathway |
| 1.1 | 24.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 1.1 | 53.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.9 | 63.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.8 | 7.0 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.7 | 25.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.7 | 18.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.7 | 28.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.6 | 31.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.6 | 25.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.6 | 7.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.6 | 5.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.5 | 80.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.5 | 2.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.5 | 6.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.5 | 8.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.5 | 26.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.5 | 15.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.5 | 2.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.4 | 10.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.4 | 5.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.4 | 3.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.4 | 12.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 9.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 11.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 5.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 2.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.3 | 7.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 5.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.3 | 8.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.3 | 25.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.3 | 5.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 2.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.3 | 8.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 4.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 3.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 10.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 10.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 2.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 4.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 5.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 7.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 1.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 4.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 3.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 11.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 7.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 10.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.2 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 0.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 2.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 2.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 7.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 112.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 7.8 | 156.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 7.5 | 120.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 7.1 | 21.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 5.8 | 86.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 4.8 | 138.0 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 3.9 | 85.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 3.5 | 112.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 3.0 | 26.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 2.8 | 35.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 2.4 | 31.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 2.3 | 107.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 2.3 | 208.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 1.9 | 45.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 1.9 | 22.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.8 | 37.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 1.8 | 14.4 | REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | Genes involved in E2F mediated regulation of DNA replication |
| 1.6 | 30.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 1.5 | 18.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 1.4 | 13.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 1.4 | 57.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 1.4 | 28.7 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 1.3 | 22.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 1.1 | 10.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 1.1 | 4.3 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 1.1 | 36.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.0 | 36.0 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 1.0 | 28.2 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 1.0 | 22.9 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 1.0 | 28.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 1.0 | 25.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.9 | 22.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.9 | 17.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.9 | 27.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.9 | 86.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.8 | 10.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.7 | 5.7 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.7 | 11.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.7 | 6.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.7 | 25.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.7 | 14.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.7 | 71.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.6 | 13.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.6 | 7.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.6 | 4.3 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.6 | 5.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.6 | 72.4 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.6 | 32.2 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.6 | 6.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.5 | 11.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.5 | 16.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.5 | 6.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.5 | 0.5 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.5 | 1.9 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.5 | 7.1 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.5 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.5 | 30.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.4 | 7.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.4 | 12.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.4 | 23.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.4 | 26.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 7.0 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.4 | 5.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 6.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.4 | 0.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 10.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.3 | 12.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.3 | 6.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.3 | 9.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 1.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.3 | 4.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.3 | 0.6 | REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | Genes involved in Signaling by TGF-beta Receptor Complex |
| 0.3 | 4.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 7.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 16.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.3 | 5.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 4.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 3.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 6.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 2.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.2 | 0.8 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.2 | 2.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 5.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 2.6 | REACTOME MITOTIC G2 G2 M PHASES | Genes involved in Mitotic G2-G2/M phases |
| 0.2 | 5.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 6.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 3.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 1.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 4.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 2.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 6.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 1.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.2 | 3.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 0.9 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 7.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.3 | REACTOME SIGNAL AMPLIFICATION | Genes involved in Signal amplification |
| 0.1 | 1.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.9 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 2.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 12.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 4.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 3.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 5.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 4.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 1.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 8.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 5.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.6 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |