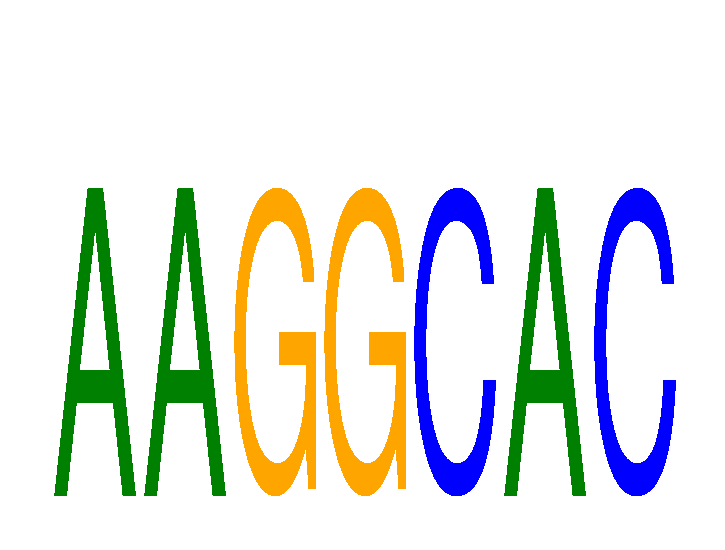Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for AAGGCAC
Z-value: 0.54
miRNA associated with seed AAGGCAC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-124-3p.1
|
MIMAT0000422 |
Activity profile of AAGGCAC motif
Sorted Z-values of AAGGCAC motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGCAC
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_132490750 | 0.90 |
ENST00000437654.6
ENST00000245414.9 ENST00000680139.1 ENST00000680352.1 ENST00000679440.1 ENST00000680903.1 |
IRF1
|
interferon regulatory factor 1 |
| chr4_-_119628791 | 0.63 |
ENST00000354960.8
|
PDE5A
|
phosphodiesterase 5A |
| chr11_-_72674394 | 0.63 |
ENST00000418754.6
ENST00000334456.10 ENST00000542969.2 |
PDE2A
|
phosphodiesterase 2A |
| chr9_+_113150991 | 0.60 |
ENST00000259392.8
|
SLC31A2
|
solute carrier family 31 member 2 |
| chr5_+_128083757 | 0.57 |
ENST00000262461.7
ENST00000628403.2 ENST00000343225.4 |
SLC12A2
|
solute carrier family 12 member 2 |
| chr8_+_17497078 | 0.56 |
ENST00000494857.6
ENST00000522656.5 |
SLC7A2
|
solute carrier family 7 member 2 |
| chr6_-_154510675 | 0.56 |
ENST00000607772.6
|
CNKSR3
|
CNKSR family member 3 |
| chr6_-_142946312 | 0.54 |
ENST00000367604.6
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr7_-_24980148 | 0.50 |
ENST00000313367.7
|
OSBPL3
|
oxysterol binding protein like 3 |
| chr21_+_25639251 | 0.50 |
ENST00000480456.6
|
JAM2
|
junctional adhesion molecule 2 |
| chr12_+_121132869 | 0.50 |
ENST00000328963.10
|
P2RX7
|
purinergic receptor P2X 7 |
| chr8_+_23528947 | 0.48 |
ENST00000519973.6
|
SLC25A37
|
solute carrier family 25 member 37 |
| chr3_-_187745460 | 0.46 |
ENST00000406870.7
|
BCL6
|
BCL6 transcription repressor |
| chr12_-_104958268 | 0.46 |
ENST00000432951.1
ENST00000258538.8 ENST00000415674.1 ENST00000424946.1 ENST00000433540.5 |
SLC41A2
|
solute carrier family 41 member 2 |
| chr14_+_21070273 | 0.44 |
ENST00000555038.5
ENST00000298694.9 |
ARHGEF40
|
Rho guanine nucleotide exchange factor 40 |
| chr3_-_165196369 | 0.43 |
ENST00000475390.2
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chrX_+_68829009 | 0.41 |
ENST00000204961.5
|
EFNB1
|
ephrin B1 |
| chr5_-_180353317 | 0.41 |
ENST00000253778.13
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr5_-_32312913 | 0.41 |
ENST00000280285.9
ENST00000382142.8 ENST00000264934.5 |
MTMR12
|
myotubularin related protein 12 |
| chr2_+_109129199 | 0.40 |
ENST00000309415.8
|
SH3RF3
|
SH3 domain containing ring finger 3 |
| chr6_+_71288803 | 0.39 |
ENST00000370435.5
|
OGFRL1
|
opioid growth factor receptor like 1 |
| chr7_-_108456378 | 0.38 |
ENST00000613830.4
ENST00000413765.6 ENST00000379028.8 |
NRCAM
|
neuronal cell adhesion molecule |
| chr8_-_37899454 | 0.38 |
ENST00000522727.5
ENST00000287263.8 ENST00000330843.9 |
RAB11FIP1
|
RAB11 family interacting protein 1 |
| chr16_-_84618067 | 0.38 |
ENST00000262428.5
|
COTL1
|
coactosin like F-actin binding protein 1 |
| chr14_+_54567612 | 0.38 |
ENST00000251091.9
ENST00000392067.7 ENST00000631086.2 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr20_-_10673987 | 0.37 |
ENST00000254958.10
|
JAG1
|
jagged canonical Notch ligand 1 |
| chr6_+_37170133 | 0.35 |
ENST00000373509.6
|
PIM1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr2_+_26346086 | 0.35 |
ENST00000613142.4
ENST00000260585.12 ENST00000447170.1 |
SELENOI
|
selenoprotein I |
| chr11_-_128522264 | 0.34 |
ENST00000531611.5
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr10_+_72692125 | 0.34 |
ENST00000373053.7
ENST00000357157.10 |
MCU
|
mitochondrial calcium uniporter |
| chr5_-_95961830 | 0.33 |
ENST00000513343.1
ENST00000237853.9 |
ELL2
|
elongation factor for RNA polymerase II 2 |
| chr16_+_12901591 | 0.31 |
ENST00000558583.3
|
SHISA9
|
shisa family member 9 |
| chr12_-_89524734 | 0.31 |
ENST00000529983.3
|
GALNT4
|
polypeptide N-acetylgalactosaminyltransferase 4 |
| chr5_-_59893718 | 0.31 |
ENST00000340635.11
|
PDE4D
|
phosphodiesterase 4D |
| chr15_-_55270383 | 0.31 |
ENST00000396307.6
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_+_112055201 | 0.30 |
ENST00000283206.9
|
TMEM87B
|
transmembrane protein 87B |
| chr15_+_22786610 | 0.30 |
ENST00000337435.9
|
NIPA1
|
NIPA magnesium transporter 1 |
| chr8_+_85177225 | 0.30 |
ENST00000418930.6
|
E2F5
|
E2F transcription factor 5 |
| chr3_+_119468952 | 0.30 |
ENST00000476573.5
ENST00000295588.9 |
POGLUT1
|
protein O-glucosyltransferase 1 |
| chr12_-_89526253 | 0.30 |
ENST00000547474.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr20_+_46894824 | 0.29 |
ENST00000327619.10
ENST00000497062.6 |
EYA2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr2_-_288056 | 0.29 |
ENST00000403610.9
|
ALKAL2
|
ALK and LTK ligand 2 |
| chr1_-_169485931 | 0.29 |
ENST00000367804.4
ENST00000646596.1 ENST00000236137.10 |
SLC19A2
|
solute carrier family 19 member 2 |
| chr12_-_57078739 | 0.29 |
ENST00000379391.7
|
NEMP1
|
nuclear envelope integral membrane protein 1 |
| chr3_-_185498964 | 0.28 |
ENST00000296254.3
|
TMEM41A
|
transmembrane protein 41A |
| chr10_+_70815889 | 0.28 |
ENST00000373202.8
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr2_-_37671633 | 0.28 |
ENST00000295324.4
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr9_+_137605674 | 0.28 |
ENST00000371421.9
ENST00000431925.6 |
ARRDC1
|
arrestin domain containing 1 |
| chr18_+_22169580 | 0.27 |
ENST00000269216.10
|
GATA6
|
GATA binding protein 6 |
| chr18_-_34224871 | 0.27 |
ENST00000261592.10
|
NOL4
|
nucleolar protein 4 |
| chr14_+_69611586 | 0.27 |
ENST00000342745.5
|
SUSD6
|
sushi domain containing 6 |
| chr1_+_100896060 | 0.26 |
ENST00000370112.8
ENST00000357650.9 |
SLC30A7
|
solute carrier family 30 member 7 |
| chr7_-_139777986 | 0.26 |
ENST00000406875.8
|
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr20_-_45348414 | 0.26 |
ENST00000372733.3
|
SDC4
|
syndecan 4 |
| chr11_-_64878612 | 0.26 |
ENST00000320631.8
|
EHD1
|
EH domain containing 1 |
| chr10_-_103855406 | 0.26 |
ENST00000355946.6
ENST00000369774.8 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr6_-_75206044 | 0.26 |
ENST00000322507.13
|
COL12A1
|
collagen type XII alpha 1 chain |
| chr3_+_105366877 | 0.25 |
ENST00000306107.9
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr16_-_67806513 | 0.25 |
ENST00000317506.8
ENST00000448631.6 ENST00000602677.5 ENST00000630626.2 |
RANBP10
|
RAN binding protein 10 |
| chr12_-_110583305 | 0.25 |
ENST00000354300.5
|
PPTC7
|
protein phosphatase targeting COQ7 |
| chr3_-_122564577 | 0.24 |
ENST00000477522.6
ENST00000360356.6 |
PARP9
|
poly(ADP-ribose) polymerase family member 9 |
| chr12_-_128823942 | 0.24 |
ENST00000266771.10
|
SLC15A4
|
solute carrier family 15 member 4 |
| chr6_-_154356735 | 0.24 |
ENST00000367220.8
ENST00000265198.8 ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr1_-_18956669 | 0.24 |
ENST00000455833.7
|
IFFO2
|
intermediate filament family orphan 2 |
| chr6_-_134318097 | 0.24 |
ENST00000367858.10
ENST00000533224.1 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr15_+_79432330 | 0.24 |
ENST00000305428.8
|
MINAR1
|
membrane integral NOTCH2 associated receptor 1 |
| chr5_+_151771884 | 0.24 |
ENST00000627077.2
ENST00000678976.1 ENST00000677408.1 ENST00000678070.1 ENST00000678964.1 ENST00000678925.1 ENST00000394123.7 ENST00000522761.6 ENST00000676827.1 |
G3BP1
|
G3BP stress granule assembly factor 1 |
| chr2_-_64653906 | 0.24 |
ENST00000313349.3
|
SERTAD2
|
SERTA domain containing 2 |
| chr8_-_63086031 | 0.23 |
ENST00000260116.5
|
TTPA
|
alpha tocopherol transfer protein |
| chr5_-_27038576 | 0.23 |
ENST00000511822.1
ENST00000231021.9 |
CDH9
|
cadherin 9 |
| chr13_-_43786889 | 0.23 |
ENST00000261488.10
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr1_-_41918858 | 0.23 |
ENST00000372583.6
|
HIVEP3
|
HIVEP zinc finger 3 |
| chr2_+_73214233 | 0.22 |
ENST00000389501.9
ENST00000629411.2 |
SMYD5
|
SMYD family member 5 |
| chr6_-_13814490 | 0.22 |
ENST00000379170.8
|
MCUR1
|
mitochondrial calcium uniporter regulator 1 |
| chr14_+_35046238 | 0.22 |
ENST00000280987.9
|
FAM177A1
|
family with sequence similarity 177 member A1 |
| chr3_-_27456743 | 0.22 |
ENST00000295736.9
ENST00000428386.5 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4 member 7 |
| chr19_+_676385 | 0.22 |
ENST00000166139.9
|
FSTL3
|
follistatin like 3 |
| chr13_-_29595670 | 0.22 |
ENST00000380752.10
|
SLC7A1
|
solute carrier family 7 member 1 |
| chr17_-_51120855 | 0.22 |
ENST00000618113.4
ENST00000357122.8 ENST00000262013.12 |
SPAG9
|
sperm associated antigen 9 |
| chr11_+_125592826 | 0.22 |
ENST00000529196.5
ENST00000392708.9 ENST00000649491.1 ENST00000531491.5 |
STT3A
|
STT3 oligosaccharyltransferase complex catalytic subunit A |
| chr5_-_124744513 | 0.21 |
ENST00000504926.5
|
ZNF608
|
zinc finger protein 608 |
| chr1_-_16156059 | 0.21 |
ENST00000358432.8
|
EPHA2
|
EPH receptor A2 |
| chr20_-_18057841 | 0.21 |
ENST00000278780.7
|
OVOL2
|
ovo like zinc finger 2 |
| chr9_+_116153783 | 0.21 |
ENST00000328252.4
|
PAPPA
|
pappalysin 1 |
| chr16_-_11586941 | 0.21 |
ENST00000571976.1
ENST00000413364.6 |
LITAF
|
lipopolysaccharide induced TNF factor |
| chr11_+_59172116 | 0.21 |
ENST00000227451.4
|
DTX4
|
deltex E3 ubiquitin ligase 4 |
| chr17_-_43778937 | 0.21 |
ENST00000226004.8
|
DUSP3
|
dual specificity phosphatase 3 |
| chr5_-_116574802 | 0.21 |
ENST00000343348.11
|
SEMA6A
|
semaphorin 6A |
| chr1_-_226737277 | 0.21 |
ENST00000272117.7
|
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr4_+_123399488 | 0.20 |
ENST00000394339.2
|
SPRY1
|
sprouty RTK signaling antagonist 1 |
| chr1_+_1020068 | 0.20 |
ENST00000379370.7
ENST00000620552.4 |
AGRN
|
agrin |
| chr4_-_80073170 | 0.20 |
ENST00000403729.7
|
ANTXR2
|
ANTXR cell adhesion molecule 2 |
| chr9_-_112333603 | 0.20 |
ENST00000450374.1
ENST00000374257.6 ENST00000374255.6 ENST00000334318.10 |
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr2_+_11746576 | 0.20 |
ENST00000256720.6
ENST00000674199.1 ENST00000441684.5 ENST00000423495.1 |
LPIN1
|
lipin 1 |
| chr12_+_63844663 | 0.20 |
ENST00000355086.8
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr20_+_380747 | 0.20 |
ENST00000217233.9
|
TRIB3
|
tribbles pseudokinase 3 |
| chr6_-_33789090 | 0.20 |
ENST00000614475.4
ENST00000293760.10 |
LEMD2
|
LEM domain nuclear envelope protein 2 |
| chr15_-_50765656 | 0.20 |
ENST00000261854.10
|
SPPL2A
|
signal peptide peptidase like 2A |
| chr19_-_3761657 | 0.19 |
ENST00000316757.4
|
APBA3
|
amyloid beta precursor protein binding family A member 3 |
| chr6_+_12012304 | 0.19 |
ENST00000379388.7
ENST00000627968.2 ENST00000541134.5 |
HIVEP1
|
HIVEP zinc finger 1 |
| chr3_-_142448060 | 0.19 |
ENST00000264951.8
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr1_+_65147514 | 0.19 |
ENST00000545314.5
|
AK4
|
adenylate kinase 4 |
| chr10_-_104338431 | 0.19 |
ENST00000647721.1
ENST00000337478.3 |
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr3_+_36380477 | 0.19 |
ENST00000457375.6
ENST00000273183.8 ENST00000434649.1 |
STAC
|
SH3 and cysteine rich domain |
| chr11_+_118606428 | 0.19 |
ENST00000361417.6
|
PHLDB1
|
pleckstrin homology like domain family B member 1 |
| chr5_-_132227808 | 0.19 |
ENST00000401867.5
ENST00000379086.5 ENST00000379100.7 ENST00000418055.5 ENST00000453286.5 ENST00000360568.8 ENST00000379104.7 ENST00000166534.8 |
P4HA2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr11_+_33257265 | 0.19 |
ENST00000303296.9
ENST00000379016.7 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr4_+_94757921 | 0.19 |
ENST00000515059.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr2_+_64989343 | 0.19 |
ENST00000234256.4
|
SLC1A4
|
solute carrier family 1 member 4 |
| chr4_-_41214602 | 0.19 |
ENST00000508676.5
ENST00000506352.5 ENST00000295974.12 |
APBB2
|
amyloid beta precursor protein binding family B member 2 |
| chr6_-_43575966 | 0.18 |
ENST00000265351.12
|
XPO5
|
exportin 5 |
| chr2_+_46941199 | 0.18 |
ENST00000319190.11
ENST00000394850.6 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr5_+_150508110 | 0.18 |
ENST00000261797.7
|
NDST1
|
N-deacetylase and N-sulfotransferase 1 |
| chr17_-_35121487 | 0.18 |
ENST00000593039.5
|
ENSG00000267618.5
|
RAD51L3-RFFL readthrough |
| chr18_-_59359245 | 0.18 |
ENST00000251047.6
|
LMAN1
|
lectin, mannose binding 1 |
| chr5_+_56815534 | 0.17 |
ENST00000399503.4
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1 |
| chr1_+_155135817 | 0.17 |
ENST00000303343.12
ENST00000368404.9 |
SLC50A1
|
solute carrier family 50 member 1 |
| chr3_-_47781837 | 0.17 |
ENST00000254480.10
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 1 |
| chr17_-_76453142 | 0.17 |
ENST00000319380.12
|
UBE2O
|
ubiquitin conjugating enzyme E2 O |
| chr3_-_64687613 | 0.17 |
ENST00000295903.8
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif 9 |
| chr4_+_47031551 | 0.17 |
ENST00000295454.8
|
GABRB1
|
gamma-aminobutyric acid type A receptor subunit beta1 |
| chr19_-_1863497 | 0.17 |
ENST00000617223.1
ENST00000250916.6 |
KLF16
|
Kruppel like factor 16 |
| chr4_+_107824555 | 0.17 |
ENST00000394684.8
|
SGMS2
|
sphingomyelin synthase 2 |
| chr1_-_243850070 | 0.16 |
ENST00000366539.6
ENST00000672578.1 |
AKT3
|
AKT serine/threonine kinase 3 |
| chr1_-_27490045 | 0.16 |
ENST00000536657.1
|
WASF2
|
WASP family member 2 |
| chr5_-_16936231 | 0.16 |
ENST00000507288.1
ENST00000274203.13 ENST00000513610.6 |
MYO10
|
myosin X |
| chr4_+_25234003 | 0.16 |
ENST00000264864.8
|
PI4K2B
|
phosphatidylinositol 4-kinase type 2 beta |
| chr20_-_46406582 | 0.16 |
ENST00000450812.5
ENST00000290246.11 ENST00000396391.5 |
ELMO2
|
engulfment and cell motility 2 |
| chr1_-_100894775 | 0.16 |
ENST00000416479.1
ENST00000370113.7 |
EXTL2
|
exostosin like glycosyltransferase 2 |
| chr1_-_197201262 | 0.16 |
ENST00000367405.5
|
ZBTB41
|
zinc finger and BTB domain containing 41 |
| chr2_-_164841410 | 0.16 |
ENST00000342193.8
ENST00000375458.6 |
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr17_-_65056659 | 0.16 |
ENST00000439174.7
|
GNA13
|
G protein subunit alpha 13 |
| chr2_+_238426920 | 0.16 |
ENST00000264607.9
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr18_+_80109236 | 0.16 |
ENST00000262198.9
ENST00000560752.5 |
ADNP2
|
ADNP homeobox 2 |
| chr12_+_123712333 | 0.16 |
ENST00000330342.8
ENST00000613625.5 |
ATP6V0A2
|
ATPase H+ transporting V0 subunit a2 |
| chr2_-_24360299 | 0.15 |
ENST00000361999.7
|
ITSN2
|
intersectin 2 |
| chr19_-_43596123 | 0.15 |
ENST00000422989.6
ENST00000598324.1 |
IRGQ
|
immunity related GTPase Q |
| chr21_-_17819386 | 0.15 |
ENST00000400559.7
ENST00000400558.7 |
C21orf91
|
chromosome 21 open reading frame 91 |
| chr13_-_25287457 | 0.15 |
ENST00000381801.6
|
MTMR6
|
myotubularin related protein 6 |
| chr10_-_73414027 | 0.15 |
ENST00000372921.10
ENST00000372919.8 |
ANXA7
|
annexin A7 |
| chr1_+_205504592 | 0.15 |
ENST00000506784.5
ENST00000360066.6 |
CDK18
|
cyclin dependent kinase 18 |
| chr6_-_107115493 | 0.15 |
ENST00000369042.6
|
BEND3
|
BEN domain containing 3 |
| chr12_-_57006476 | 0.15 |
ENST00000300101.3
|
ZBTB39
|
zinc finger and BTB domain containing 39 |
| chr10_-_58267868 | 0.14 |
ENST00000373935.4
|
IPMK
|
inositol polyphosphate multikinase |
| chr9_-_99221897 | 0.14 |
ENST00000476832.2
|
ALG2
|
ALG2 alpha-1,3/1,6-mannosyltransferase |
| chr10_-_74150781 | 0.14 |
ENST00000355264.9
ENST00000372745.1 |
AP3M1
|
adaptor related protein complex 3 subunit mu 1 |
| chr11_-_45665578 | 0.14 |
ENST00000308064.7
|
CHST1
|
carbohydrate sulfotransferase 1 |
| chr8_+_28701487 | 0.14 |
ENST00000220562.9
|
EXTL3
|
exostosin like glycosyltransferase 3 |
| chr12_+_27780224 | 0.14 |
ENST00000381271.7
|
KLHL42
|
kelch like family member 42 |
| chr15_-_72117712 | 0.14 |
ENST00000444904.5
ENST00000564571.5 |
MYO9A
|
myosin IXA |
| chr12_-_99154492 | 0.14 |
ENST00000546568.5
ENST00000546960.5 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr3_-_154324446 | 0.14 |
ENST00000308361.10
ENST00000496811.6 |
DHX36
|
DEAH-box helicase 36 |
| chr14_-_76812804 | 0.14 |
ENST00000556298.1
ENST00000251089.8 |
ANGEL1
|
angel homolog 1 |
| chr4_+_76949743 | 0.14 |
ENST00000502584.5
ENST00000264893.11 ENST00000510641.5 |
SEPTIN11
|
septin 11 |
| chr1_-_55215345 | 0.14 |
ENST00000294383.7
|
USP24
|
ubiquitin specific peptidase 24 |
| chr6_+_30326835 | 0.14 |
ENST00000440271.5
ENST00000376656.8 ENST00000396551.7 ENST00000428728.5 ENST00000396548.5 ENST00000428404.5 |
TRIM39
|
tripartite motif containing 39 |
| chr19_+_17555615 | 0.14 |
ENST00000252599.9
|
COLGALT1
|
collagen beta(1-O)galactosyltransferase 1 |
| chr2_-_11344580 | 0.14 |
ENST00000616279.4
ENST00000315872.11 |
ROCK2
|
Rho associated coiled-coil containing protein kinase 2 |
| chr14_-_44961889 | 0.14 |
ENST00000579157.1
ENST00000396128.9 ENST00000556500.1 |
KLHL28
|
kelch like family member 28 |
| chr16_-_48610150 | 0.14 |
ENST00000262384.4
|
N4BP1
|
NEDD4 binding protein 1 |
| chr3_+_191329342 | 0.14 |
ENST00000392455.9
|
CCDC50
|
coiled-coil domain containing 50 |
| chr8_-_544825 | 0.14 |
ENST00000324079.11
|
TDRP
|
testis development related protein |
| chr13_+_42048645 | 0.14 |
ENST00000337343.9
ENST00000261491.9 ENST00000611224.1 |
DGKH
|
diacylglycerol kinase eta |
| chr9_+_97501622 | 0.14 |
ENST00000259365.9
|
TMOD1
|
tropomodulin 1 |
| chrX_+_104166436 | 0.13 |
ENST00000493442.2
|
FAM199X
|
family with sequence similarity 199, X-linked |
| chr20_+_34704336 | 0.13 |
ENST00000374809.6
ENST00000374810.8 ENST00000451665.5 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr2_+_46542474 | 0.13 |
ENST00000238738.9
|
RHOQ
|
ras homolog family member Q |
| chr14_-_54902807 | 0.13 |
ENST00000543643.6
ENST00000536224.2 ENST00000395514.5 ENST00000491895.7 |
GCH1
|
GTP cyclohydrolase 1 |
| chr4_-_88697810 | 0.13 |
ENST00000323061.7
|
NAP1L5
|
nucleosome assembly protein 1 like 5 |
| chr17_-_17591658 | 0.13 |
ENST00000435340.6
ENST00000255389.10 ENST00000395781.6 |
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr1_+_168280872 | 0.13 |
ENST00000367821.8
|
TBX19
|
T-box transcription factor 19 |
| chr12_-_389249 | 0.13 |
ENST00000535014.5
ENST00000543507.1 ENST00000544760.1 ENST00000399788.7 |
KDM5A
|
lysine demethylase 5A |
| chr4_+_48341505 | 0.13 |
ENST00000264313.11
|
SLAIN2
|
SLAIN motif family member 2 |
| chr3_-_120094436 | 0.13 |
ENST00000264235.13
ENST00000677034.1 |
GSK3B
|
glycogen synthase kinase 3 beta |
| chr11_-_115504389 | 0.13 |
ENST00000545380.5
ENST00000452722.7 ENST00000331581.11 ENST00000537058.5 ENST00000536727.5 ENST00000542447.6 |
CADM1
|
cell adhesion molecule 1 |
| chr20_+_49936371 | 0.13 |
ENST00000244061.6
ENST00000625177.3 ENST00000622920.1 |
RNF114
|
ring finger protein 114 |
| chr1_+_183023409 | 0.13 |
ENST00000258341.5
|
LAMC1
|
laminin subunit gamma 1 |
| chr19_+_38647614 | 0.13 |
ENST00000252699.7
ENST00000424234.7 ENST00000440400.2 |
ACTN4
|
actinin alpha 4 |
| chr8_-_15238423 | 0.13 |
ENST00000382080.6
|
SGCZ
|
sarcoglycan zeta |
| chr1_+_113929600 | 0.13 |
ENST00000369558.5
ENST00000369561.8 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr4_-_99563984 | 0.13 |
ENST00000394877.7
|
TRMT10A
|
tRNA methyltransferase 10A |
| chr5_+_31639104 | 0.13 |
ENST00000438447.2
|
PDZD2
|
PDZ domain containing 2 |
| chr9_-_35732122 | 0.13 |
ENST00000314888.10
|
TLN1
|
talin 1 |
| chr14_-_103562637 | 0.13 |
ENST00000299204.6
|
BAG5
|
BAG cochaperone 5 |
| chrX_+_154458274 | 0.13 |
ENST00000369682.4
|
PLXNA3
|
plexin A3 |
| chr2_+_72917489 | 0.13 |
ENST00000258106.11
|
EMX1
|
empty spiracles homeobox 1 |
| chr22_-_36387949 | 0.12 |
ENST00000216181.11
|
MYH9
|
myosin heavy chain 9 |
| chr10_-_27154226 | 0.12 |
ENST00000427324.5
ENST00000375972.7 ENST00000326799.7 |
YME1L1
|
YME1 like 1 ATPase |
| chr22_+_21642287 | 0.12 |
ENST00000248958.5
|
SDF2L1
|
stromal cell derived factor 2 like 1 |
| chr3_+_153162196 | 0.12 |
ENST00000323534.5
|
RAP2B
|
RAP2B, member of RAS oncogene family |
| chr1_+_77779618 | 0.12 |
ENST00000370791.7
ENST00000443751.3 ENST00000645756.1 ENST00000643390.1 ENST00000642959.1 |
MIGA1
|
mitoguardin 1 |
| chr11_+_111602380 | 0.12 |
ENST00000304987.4
|
SIK2
|
salt inducible kinase 2 |
| chr17_-_1179940 | 0.12 |
ENST00000302538.10
|
ABR
|
ABR activator of RhoGEF and GTPase |
| chr3_+_50236192 | 0.12 |
ENST00000313601.11
|
GNAI2
|
G protein subunit alpha i2 |
| chr1_-_37554277 | 0.12 |
ENST00000296215.8
|
SNIP1
|
Smad nuclear interacting protein 1 |
| chr15_+_45587366 | 0.12 |
ENST00000220531.9
|
BLOC1S6
|
biogenesis of lysosomal organelles complex 1 subunit 6 |
| chr1_-_222712428 | 0.12 |
ENST00000355727.3
ENST00000340020.11 |
AIDA
|
axin interactor, dorsalization associated |
| chr12_+_4273751 | 0.12 |
ENST00000675880.1
ENST00000261254.8 |
CCND2
|
cyclin D2 |
| chr21_+_43719095 | 0.12 |
ENST00000468090.5
ENST00000291565.9 |
PDXK
|
pyridoxal kinase |
| chr10_+_69451456 | 0.12 |
ENST00000373290.7
|
TSPAN15
|
tetraspanin 15 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.2 | 0.6 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 0.6 | GO:1903400 | L-arginine transmembrane transport(GO:1903400) |
| 0.2 | 0.5 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.2 | 0.5 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.6 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.5 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.4 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.1 | GO:0001743 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.3 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.1 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.3 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.1 | 0.3 | GO:0070378 | positive regulation of ERK5 cascade(GO:0070378) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.3 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.2 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.2 | GO:0042946 | glucoside transport(GO:0042946) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.6 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 0.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.1 | 0.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.6 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:0042360 | vitamin E metabolic process(GO:0042360) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 0.3 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.3 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.3 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.2 | GO:1903921 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.0 | 0.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.8 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.4 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.1 | GO:2000077 | negative regulation of dopaminergic neuron differentiation(GO:1904339) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.1 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0003343 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.0 | 0.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.1 | GO:0051463 | negative regulation of cortisol secretion(GO:0051463) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:1902594 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 0.0 | 0.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.1 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.5 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.0 | 0.2 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.0 | GO:1903179 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0042853 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0090176 | microtubule cytoskeleton organization involved in establishment of planar polarity(GO:0090176) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.0 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.0 | 0.1 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.0 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.0 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.1 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.0 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.0 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 0.2 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.5 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0098560 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 0.6 | GO:0005292 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.1 | 0.4 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.3 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.2 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 0.3 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 0.3 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 0.2 | GO:0042947 | glucoside transmembrane transporter activity(GO:0042947) |
| 0.1 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.6 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.5 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.2 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.4 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.7 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.0 | GO:0031753 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.0 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |