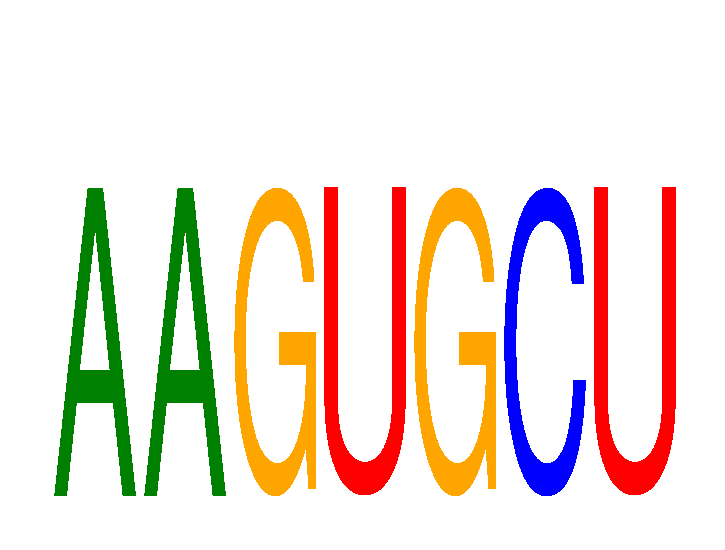Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for AAGUGCU
Z-value: 0.40
miRNA associated with seed AAGUGCU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-302a-3p
|
MIMAT0000684 |
|
hsa-miR-302b-3p
|
MIMAT0000715 |
|
hsa-miR-302c-3p.1
|
MIMAT0000717 |
|
hsa-miR-302d-3p
|
MIMAT0000718 |
|
hsa-miR-302e
|
MIMAT0005931 |
|
hsa-miR-372-3p
|
MIMAT0000724 |
|
hsa-miR-373-3p
|
MIMAT0000726 |
|
hsa-miR-520a-3p
|
MIMAT0002834 |
|
hsa-miR-520b
|
MIMAT0002843 |
|
hsa-miR-520c-3p
|
MIMAT0002846 |
|
hsa-miR-520d-3p
|
MIMAT0002856 |
|
hsa-miR-520e
|
MIMAT0002825 |
Activity profile of AAGUGCU motif
Sorted Z-values of AAGUGCU motif
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGUGCU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_155667198 | 1.44 |
ENST00000296518.11
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr4_+_125314918 | 1.21 |
ENST00000674496.2
ENST00000394329.9 |
FAT4
|
FAT atypical cadherin 4 |
| chr2_-_189580773 | 1.19 |
ENST00000261024.7
|
SLC40A1
|
solute carrier family 40 member 1 |
| chr9_-_14314067 | 0.97 |
ENST00000397575.7
|
NFIB
|
nuclear factor I B |
| chr10_-_79445617 | 0.90 |
ENST00000372336.4
|
ZCCHC24
|
zinc finger CCHC-type containing 24 |
| chr6_-_16761447 | 0.85 |
ENST00000244769.8
ENST00000436367.6 |
ATXN1
|
ataxin 1 |
| chr12_+_27244222 | 0.84 |
ENST00000545470.5
ENST00000389032.8 ENST00000540996.5 |
STK38L
|
serine/threonine kinase 38 like |
| chr3_+_20040437 | 0.83 |
ENST00000263754.5
|
KAT2B
|
lysine acetyltransferase 2B |
| chr9_-_131270493 | 0.78 |
ENST00000372269.7
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78 member A |
| chr10_+_110871903 | 0.72 |
ENST00000280154.12
|
PDCD4
|
programmed cell death 4 |
| chr1_+_61082553 | 0.68 |
ENST00000403491.8
ENST00000371187.7 |
NFIA
|
nuclear factor I A |
| chr6_+_11537738 | 0.65 |
ENST00000379426.2
|
TMEM170B
|
transmembrane protein 170B |
| chr12_+_32502114 | 0.65 |
ENST00000682739.1
ENST00000427716.7 ENST00000583694.2 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr9_-_10612966 | 0.56 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D |
| chr4_+_6269831 | 0.56 |
ENST00000503569.5
ENST00000673991.1 ENST00000682275.1 ENST00000226760.5 |
WFS1
|
wolframin ER transmembrane glycoprotein |
| chr3_+_43286512 | 0.54 |
ENST00000454177.5
ENST00000429705.6 ENST00000296088.12 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr9_+_112750722 | 0.54 |
ENST00000374232.8
|
SNX30
|
sorting nexin family member 30 |
| chr2_-_239400949 | 0.53 |
ENST00000345617.7
|
HDAC4
|
histone deacetylase 4 |
| chr12_-_31591129 | 0.53 |
ENST00000389082.10
|
DENND5B
|
DENN domain containing 5B |
| chr12_-_95790755 | 0.53 |
ENST00000343702.9
ENST00000344911.8 |
NTN4
|
netrin 4 |
| chr14_+_52552830 | 0.44 |
ENST00000321662.11
|
GPR137C
|
G protein-coupled receptor 137C |
| chr15_+_96330691 | 0.44 |
ENST00000394166.8
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr9_+_36036899 | 0.43 |
ENST00000377966.4
|
RECK
|
reversion inducing cysteine rich protein with kazal motifs |
| chr9_-_16870662 | 0.42 |
ENST00000380672.9
|
BNC2
|
basonuclin 2 |
| chrX_+_28587411 | 0.41 |
ENST00000378993.6
|
IL1RAPL1
|
interleukin 1 receptor accessory protein like 1 |
| chr1_-_234609445 | 0.40 |
ENST00000366610.7
|
IRF2BP2
|
interferon regulatory factor 2 binding protein 2 |
| chr2_-_43226594 | 0.40 |
ENST00000282388.4
|
ZFP36L2
|
ZFP36 ring finger protein like 2 |
| chr6_-_110815408 | 0.40 |
ENST00000368911.8
|
CDK19
|
cyclin dependent kinase 19 |
| chr12_+_131894615 | 0.39 |
ENST00000321867.6
|
ULK1
|
unc-51 like autophagy activating kinase 1 |
| chr13_+_39038292 | 0.38 |
ENST00000470258.5
|
NHLRC3
|
NHL repeat containing 3 |
| chr1_+_210232776 | 0.37 |
ENST00000367012.4
|
SERTAD4
|
SERTA domain containing 4 |
| chr2_+_46297397 | 0.36 |
ENST00000263734.5
|
EPAS1
|
endothelial PAS domain protein 1 |
| chr1_+_78004930 | 0.35 |
ENST00000370763.6
|
DNAJB4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr4_+_88007624 | 0.35 |
ENST00000237596.7
|
PKD2
|
polycystin 2, transient receptor potential cation channel |
| chr12_-_56189548 | 0.34 |
ENST00000347471.8
ENST00000267064.8 ENST00000394023.7 |
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 2 |
| chr1_-_20486197 | 0.33 |
ENST00000375078.4
|
CAMK2N1
|
calcium/calmodulin dependent protein kinase II inhibitor 1 |
| chr12_-_24949026 | 0.33 |
ENST00000539780.5
ENST00000546285.1 ENST00000342945.9 ENST00000261192.12 |
BCAT1
|
branched chain amino acid transaminase 1 |
| chr1_-_32702736 | 0.33 |
ENST00000373484.4
ENST00000409190.8 |
SYNC
|
syncoilin, intermediate filament protein |
| chr1_+_33256479 | 0.32 |
ENST00000539719.6
ENST00000483388.5 |
ZNF362
|
zinc finger protein 362 |
| chr1_-_227947924 | 0.31 |
ENST00000272164.6
|
WNT9A
|
Wnt family member 9A |
| chr4_-_162163989 | 0.30 |
ENST00000306100.10
ENST00000427802.2 |
FSTL5
|
follistatin like 5 |
| chr10_+_96043394 | 0.30 |
ENST00000403870.7
ENST00000265992.9 ENST00000465148.3 |
CCNJ
|
cyclin J |
| chr6_-_108074703 | 0.30 |
ENST00000193322.8
|
OSTM1
|
osteoclastogenesis associated transmembrane protein 1 |
| chr17_-_49764123 | 0.30 |
ENST00000240364.7
ENST00000506156.1 |
FAM117A
|
family with sequence similarity 117 member A |
| chr3_-_18425295 | 0.27 |
ENST00000338745.11
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr12_+_56521798 | 0.27 |
ENST00000262031.10
|
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr10_-_45535346 | 0.26 |
ENST00000453424.7
ENST00000395769.6 |
MARCHF8
|
membrane associated ring-CH-type finger 8 |
| chr1_+_84078043 | 0.26 |
ENST00000370689.6
ENST00000370688.7 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr9_-_96418334 | 0.25 |
ENST00000375256.5
|
ZNF367
|
zinc finger protein 367 |
| chr3_-_171460368 | 0.24 |
ENST00000436636.7
ENST00000465393.1 ENST00000341852.10 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr14_-_75126964 | 0.24 |
ENST00000678037.1
ENST00000553823.6 ENST00000678531.1 ENST00000238616.10 |
NEK9
|
NIMA related kinase 9 |
| chr21_-_26170654 | 0.24 |
ENST00000439274.6
ENST00000358918.7 ENST00000354192.7 ENST00000348990.9 ENST00000346798.8 ENST00000357903.7 |
APP
|
amyloid beta precursor protein |
| chr2_+_15940537 | 0.24 |
ENST00000281043.4
ENST00000638417.1 |
MYCN
|
MYCN proto-oncogene, bHLH transcription factor |
| chr12_+_82686889 | 0.23 |
ENST00000321196.8
|
TMTC2
|
transmembrane O-mannosyltransferase targeting cadherins 2 |
| chr1_-_44031446 | 0.23 |
ENST00000372310.8
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr18_-_21704763 | 0.23 |
ENST00000580981.5
ENST00000289119.7 |
ABHD3
|
abhydrolase domain containing 3, phospholipase |
| chr16_+_69187125 | 0.23 |
ENST00000336278.8
|
SNTB2
|
syntrophin beta 2 |
| chr20_-_37527723 | 0.23 |
ENST00000397135.1
ENST00000397137.5 |
BLCAP
|
BLCAP apoptosis inducing factor |
| chr13_+_57631735 | 0.22 |
ENST00000377918.8
|
PCDH17
|
protocadherin 17 |
| chr6_-_98947911 | 0.22 |
ENST00000369244.7
ENST00000229971.2 |
FBXL4
|
F-box and leucine rich repeat protein 4 |
| chr2_-_55419565 | 0.22 |
ENST00000647341.1
ENST00000647401.1 ENST00000336838.10 ENST00000621814.4 ENST00000644033.1 ENST00000645477.1 ENST00000647517.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr1_+_32741779 | 0.22 |
ENST00000401073.7
|
KIAA1522
|
KIAA1522 |
| chr3_-_69386079 | 0.22 |
ENST00000398540.8
|
FRMD4B
|
FERM domain containing 4B |
| chr11_+_73308237 | 0.22 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17 |
| chr9_-_104928139 | 0.22 |
ENST00000423487.6
ENST00000374733.1 ENST00000374736.8 ENST00000678995.1 |
ABCA1
|
ATP binding cassette subfamily A member 1 |
| chr10_+_76318330 | 0.21 |
ENST00000496424.2
|
LRMDA
|
leucine rich melanocyte differentiation associated |
| chr10_+_68560317 | 0.21 |
ENST00000373644.5
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr1_-_156082412 | 0.20 |
ENST00000532414.3
|
MEX3A
|
mex-3 RNA binding family member A |
| chr7_+_87345656 | 0.20 |
ENST00000331536.8
ENST00000419147.6 ENST00000412227.6 |
CROT
|
carnitine O-octanoyltransferase |
| chr1_+_28369705 | 0.19 |
ENST00000373839.8
|
PHACTR4
|
phosphatase and actin regulator 4 |
| chr19_-_31349408 | 0.18 |
ENST00000240587.5
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr11_-_65614195 | 0.18 |
ENST00000309100.8
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr7_+_107168961 | 0.18 |
ENST00000468410.5
ENST00000478930.5 ENST00000464009.1 ENST00000222574.9 |
HBP1
|
HMG-box transcription factor 1 |
| chr8_-_123396412 | 0.18 |
ENST00000287394.10
|
ATAD2
|
ATPase family AAA domain containing 2 |
| chr2_+_197804583 | 0.18 |
ENST00000428675.6
|
PLCL1
|
phospholipase C like 1 (inactive) |
| chr9_+_125747345 | 0.18 |
ENST00000342287.9
ENST00000373489.10 ENST00000373487.8 |
PBX3
|
PBX homeobox 3 |
| chr1_+_213987929 | 0.17 |
ENST00000498508.6
ENST00000366958.9 |
PROX1
|
prospero homeobox 1 |
| chr2_-_217944005 | 0.17 |
ENST00000611415.4
ENST00000615025.5 ENST00000449814.1 ENST00000171887.8 |
TNS1
|
tensin 1 |
| chr18_-_31684504 | 0.17 |
ENST00000383131.3
ENST00000237019.11 ENST00000306851.10 |
B4GALT6
|
beta-1,4-galactosyltransferase 6 |
| chrX_-_20266834 | 0.17 |
ENST00000379565.9
|
RPS6KA3
|
ribosomal protein S6 kinase A3 |
| chr11_-_73598183 | 0.17 |
ENST00000064778.8
|
FAM168A
|
family with sequence similarity 168 member A |
| chr5_-_149551381 | 0.16 |
ENST00000670598.1
ENST00000657001.1 ENST00000515768.6 ENST00000261798.10 ENST00000377843.8 |
CSNK1A1
|
casein kinase 1 alpha 1 |
| chr17_+_40121955 | 0.16 |
ENST00000398532.9
|
MSL1
|
MSL complex subunit 1 |
| chr2_-_196171565 | 0.16 |
ENST00000263955.9
|
STK17B
|
serine/threonine kinase 17b |
| chr10_+_61901678 | 0.16 |
ENST00000644638.1
ENST00000681100.1 ENST00000279873.12 |
ARID5B
|
AT-rich interaction domain 5B |
| chr10_+_100535927 | 0.16 |
ENST00000299163.7
|
HIF1AN
|
hypoxia inducible factor 1 subunit alpha inhibitor |
| chr12_-_64752871 | 0.16 |
ENST00000418919.6
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr7_+_139341311 | 0.16 |
ENST00000297534.7
ENST00000541515.3 |
FMC1
FMC1-LUC7L2
|
formation of mitochondrial complex V assembly factor 1 homolog FMC1-LUC7L2 readthrough |
| chr9_+_79571767 | 0.16 |
ENST00000376544.7
|
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr9_-_125189721 | 0.16 |
ENST00000456642.1
ENST00000373547.9 ENST00000415905.5 ENST00000451402.5 |
PPP6C
|
protein phosphatase 6 catalytic subunit |
| chr11_-_102452758 | 0.16 |
ENST00000398136.7
ENST00000361236.7 |
TMEM123
|
transmembrane protein 123 |
| chr10_-_121927979 | 0.15 |
ENST00000369040.8
ENST00000369043.8 ENST00000224652.11 |
ATE1
|
arginyltransferase 1 |
| chr3_-_149971109 | 0.15 |
ENST00000239940.11
|
PFN2
|
profilin 2 |
| chr10_+_3067496 | 0.15 |
ENST00000381125.9
|
PFKP
|
phosphofructokinase, platelet |
| chr12_+_4909895 | 0.15 |
ENST00000638821.1
ENST00000382545.5 |
ENSG00000256654.4
KCNA1
|
novel transcript, sense overlapping KCNA1 potassium voltage-gated channel subfamily A member 1 |
| chr3_-_115071333 | 0.15 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr10_+_110207587 | 0.15 |
ENST00000332674.9
ENST00000453116.5 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr17_-_68291116 | 0.15 |
ENST00000327268.8
ENST00000580666.6 |
SLC16A6
|
solute carrier family 16 member 6 |
| chr11_-_27472698 | 0.15 |
ENST00000389858.4
ENST00000379214.9 |
LGR4
|
leucine rich repeat containing G protein-coupled receptor 4 |
| chr15_-_52191387 | 0.15 |
ENST00000261837.12
|
GNB5
|
G protein subunit beta 5 |
| chr8_-_27772585 | 0.15 |
ENST00000522915.5
ENST00000356537.9 |
CCDC25
|
coiled-coil domain containing 25 |
| chr13_-_28495079 | 0.14 |
ENST00000615840.4
ENST00000282397.9 ENST00000541932.5 ENST00000539099.1 ENST00000639477.1 |
FLT1
|
fms related receptor tyrosine kinase 1 |
| chr13_+_77697679 | 0.14 |
ENST00000418532.6
|
SLAIN1
|
SLAIN motif family member 1 |
| chr17_+_49788672 | 0.14 |
ENST00000454930.6
ENST00000259021.9 ENST00000509773.5 ENST00000510819.5 ENST00000424009.6 |
KAT7
|
lysine acetyltransferase 7 |
| chr3_+_156674579 | 0.14 |
ENST00000295924.12
|
TIPARP
|
TCDD inducible poly(ADP-ribose) polymerase |
| chr1_+_26529745 | 0.14 |
ENST00000374168.7
ENST00000374166.8 |
RPS6KA1
|
ribosomal protein S6 kinase A1 |
| chr2_+_177392734 | 0.14 |
ENST00000680770.1
ENST00000637633.2 ENST00000679459.1 ENST00000409888.1 ENST00000264167.11 ENST00000642466.2 |
AGPS
|
alkylglycerone phosphate synthase |
| chr3_+_61561561 | 0.14 |
ENST00000474889.6
|
PTPRG
|
protein tyrosine phosphatase receptor type G |
| chr9_-_122227525 | 0.14 |
ENST00000373755.6
ENST00000373754.6 |
LHX6
|
LIM homeobox 6 |
| chr14_-_34713788 | 0.13 |
ENST00000341223.8
|
CFL2
|
cofilin 2 |
| chr1_-_84690406 | 0.13 |
ENST00000605755.5
ENST00000342203.8 ENST00000437941.6 |
SSX2IP
|
SSX family member 2 interacting protein |
| chr10_+_102644462 | 0.13 |
ENST00000643721.2
ENST00000302424.12 |
TRIM8
|
tripartite motif containing 8 |
| chr6_+_118894144 | 0.13 |
ENST00000229595.6
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr17_+_29593118 | 0.12 |
ENST00000394859.8
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr20_-_63969890 | 0.12 |
ENST00000369888.6
|
ZNF512B
|
zinc finger protein 512B |
| chr6_-_105137147 | 0.12 |
ENST00000314641.10
|
BVES
|
blood vessel epicardial substance |
| chr8_-_90646074 | 0.12 |
ENST00000458549.7
|
TMEM64
|
transmembrane protein 64 |
| chr1_-_94237562 | 0.12 |
ENST00000260526.11
ENST00000370217.3 |
ARHGAP29
|
Rho GTPase activating protein 29 |
| chr8_-_80874771 | 0.12 |
ENST00000327835.7
|
ZNF704
|
zinc finger protein 704 |
| chr2_+_69829630 | 0.12 |
ENST00000282570.4
|
GMCL1
|
germ cell-less 1, spermatogenesis associated |
| chr17_-_45490696 | 0.12 |
ENST00000430334.8
ENST00000584420.1 ENST00000589780.5 |
PLEKHM1
|
pleckstrin homology and RUN domain containing M1 |
| chr10_+_52314272 | 0.11 |
ENST00000373970.4
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr2_-_165794190 | 0.11 |
ENST00000392701.8
ENST00000422973.1 |
GALNT3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr11_-_40294089 | 0.11 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr1_-_153922901 | 0.11 |
ENST00000634401.1
ENST00000368655.5 |
GATAD2B
|
GATA zinc finger domain containing 2B |
| chr11_+_61752603 | 0.11 |
ENST00000278836.10
|
MYRF
|
myelin regulatory factor |
| chr2_-_20225123 | 0.11 |
ENST00000254351.9
|
SDC1
|
syndecan 1 |
| chr6_+_87155537 | 0.11 |
ENST00000369577.8
ENST00000518845.1 ENST00000339907.8 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr16_+_24539536 | 0.11 |
ENST00000568015.5
ENST00000319715.10 |
RBBP6
|
RB binding protein 6, ubiquitin ligase |
| chr7_+_100015588 | 0.10 |
ENST00000324306.11
ENST00000426572.5 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr4_+_74308463 | 0.10 |
ENST00000413830.6
|
EPGN
|
epithelial mitogen |
| chr1_-_161069666 | 0.10 |
ENST00000368016.7
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr2_+_134120169 | 0.10 |
ENST00000409645.5
|
MGAT5
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase |
| chr4_-_16226460 | 0.10 |
ENST00000405303.7
|
TAPT1
|
transmembrane anterior posterior transformation 1 |
| chr17_+_59331633 | 0.10 |
ENST00000312655.9
|
YPEL2
|
yippee like 2 |
| chr17_-_55722857 | 0.10 |
ENST00000424486.3
|
TMEM100
|
transmembrane protein 100 |
| chr17_+_46851580 | 0.10 |
ENST00000290015.7
ENST00000393461.2 |
WNT9B
|
Wnt family member 9B |
| chr19_+_13906190 | 0.10 |
ENST00000318003.11
|
CC2D1A
|
coiled-coil and C2 domain containing 1A |
| chr5_+_52989314 | 0.10 |
ENST00000296585.10
|
ITGA2
|
integrin subunit alpha 2 |
| chr7_+_20330893 | 0.10 |
ENST00000222573.5
|
ITGB8
|
integrin subunit beta 8 |
| chr13_+_114281577 | 0.10 |
ENST00000375299.8
ENST00000492270.1 ENST00000351487.5 |
UPF3A
|
UPF3A regulator of nonsense mediated mRNA decay |
| chr3_-_56801939 | 0.10 |
ENST00000296315.8
ENST00000495373.5 |
ARHGEF3
|
Rho guanine nucleotide exchange factor 3 |
| chr2_+_169827432 | 0.09 |
ENST00000272793.11
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr19_+_41262480 | 0.09 |
ENST00000352456.7
ENST00000595018.5 ENST00000597725.5 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U like 1 |
| chr14_+_56579782 | 0.09 |
ENST00000261556.11
ENST00000538838.5 |
TMEM260
|
transmembrane protein 260 |
| chr1_-_70205531 | 0.09 |
ENST00000370952.4
|
LRRC40
|
leucine rich repeat containing 40 |
| chr3_-_125375249 | 0.09 |
ENST00000484491.5
ENST00000492394.5 ENST00000471196.1 ENST00000468369.5 ENST00000485866.5 ENST00000360647.9 |
ZNF148
|
zinc finger protein 148 |
| chr1_-_231422261 | 0.09 |
ENST00000366641.4
|
EGLN1
|
egl-9 family hypoxia inducible factor 1 |
| chr7_-_127392687 | 0.09 |
ENST00000393313.5
ENST00000619291.4 ENST00000265827.8 ENST00000434602.5 |
ZNF800
|
zinc finger protein 800 |
| chr8_-_52714414 | 0.09 |
ENST00000435644.6
ENST00000518710.5 ENST00000025008.10 ENST00000517963.1 |
RB1CC1
|
RB1 inducible coiled-coil 1 |
| chrX_-_24027186 | 0.09 |
ENST00000328046.8
|
KLHL15
|
kelch like family member 15 |
| chr3_+_122795039 | 0.09 |
ENST00000261038.6
|
SLC49A4
|
solute carrier family 49 member 4 |
| chr1_+_220528112 | 0.09 |
ENST00000366917.6
ENST00000402574.5 ENST00000611084.4 ENST00000366918.8 |
MARK1
|
microtubule affinity regulating kinase 1 |
| chr9_+_126805003 | 0.09 |
ENST00000449886.5
ENST00000450858.1 ENST00000373464.5 |
ZBTB43
|
zinc finger and BTB domain containing 43 |
| chr12_+_19439469 | 0.08 |
ENST00000266508.14
|
AEBP2
|
AE binding protein 2 |
| chr7_-_105876575 | 0.08 |
ENST00000318724.8
ENST00000419735.8 |
ATXN7L1
|
ataxin 7 like 1 |
| chr11_+_22338333 | 0.08 |
ENST00000263160.4
|
SLC17A6
|
solute carrier family 17 member 6 |
| chr8_-_59119121 | 0.08 |
ENST00000361421.2
|
TOX
|
thymocyte selection associated high mobility group box |
| chr6_-_41039202 | 0.08 |
ENST00000244565.8
|
UNC5CL
|
unc-5 family C-terminal like |
| chr15_+_75843438 | 0.08 |
ENST00000267938.9
|
UBE2Q2
|
ubiquitin conjugating enzyme E2 Q2 |
| chrX_-_84188148 | 0.08 |
ENST00000262752.5
|
RPS6KA6
|
ribosomal protein S6 kinase A6 |
| chr1_-_47231715 | 0.08 |
ENST00000371884.6
|
TAL1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr17_-_49678074 | 0.08 |
ENST00000505581.5
ENST00000504102.6 ENST00000514121.6 ENST00000393328.6 ENST00000509079.6 ENST00000347630.6 |
SPOP
|
speckle type BTB/POZ protein |
| chr16_-_77435006 | 0.08 |
ENST00000282849.10
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif 18 |
| chr4_+_44678412 | 0.08 |
ENST00000281543.6
|
GUF1
|
GTP binding elongation factor GUF1 |
| chr19_-_17075418 | 0.08 |
ENST00000253669.10
|
HAUS8
|
HAUS augmin like complex subunit 8 |
| chr22_+_32801697 | 0.07 |
ENST00000266085.7
|
TIMP3
|
TIMP metallopeptidase inhibitor 3 |
| chr20_+_1894145 | 0.07 |
ENST00000400068.7
|
SIRPA
|
signal regulatory protein alpha |
| chr1_+_209827964 | 0.07 |
ENST00000491415.7
|
UTP25
|
UTP25 small subunit processor component |
| chr1_-_225889143 | 0.07 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr5_-_83077343 | 0.07 |
ENST00000502346.2
|
TMEM167A
|
transmembrane protein 167A |
| chr15_+_31326807 | 0.07 |
ENST00000307145.4
|
KLF13
|
Kruppel like factor 13 |
| chr11_+_118530990 | 0.07 |
ENST00000411589.6
ENST00000359862.8 ENST00000442938.6 |
TMEM25
|
transmembrane protein 25 |
| chr13_-_25172278 | 0.07 |
ENST00000515384.2
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr9_+_117704382 | 0.07 |
ENST00000646089.1
ENST00000355622.8 |
ENSG00000285082.2
TLR4
|
novel protein toll like receptor 4 |
| chr2_+_96816236 | 0.07 |
ENST00000377060.7
ENST00000305510.4 |
CNNM3
|
cyclin and CBS domain divalent metal cation transport mediator 3 |
| chr3_+_58306236 | 0.07 |
ENST00000295959.10
ENST00000445193.7 ENST00000466547.1 ENST00000475412.5 ENST00000474660.5 ENST00000477305.5 ENST00000481972.5 |
RPP14
HTD2
|
ribonuclease P/MRP subunit p14 hydroxyacyl-thioester dehydratase type 2 |
| chr6_+_20401864 | 0.07 |
ENST00000346618.8
ENST00000613242.4 |
E2F3
|
E2F transcription factor 3 |
| chr1_-_53328053 | 0.07 |
ENST00000371454.6
ENST00000667377.1 ENST00000306052.12 ENST00000668448.1 |
LRP8
|
LDL receptor related protein 8 |
| chr10_+_63521365 | 0.07 |
ENST00000373758.5
|
REEP3
|
receptor accessory protein 3 |
| chr1_-_23369813 | 0.07 |
ENST00000314011.9
|
ZNF436
|
zinc finger protein 436 |
| chr4_-_124712721 | 0.07 |
ENST00000504087.6
ENST00000515641.1 |
ANKRD50
|
ankyrin repeat domain 50 |
| chr15_-_78234513 | 0.06 |
ENST00000558130.1
ENST00000258873.9 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr1_+_86914616 | 0.06 |
ENST00000370550.10
ENST00000370551.8 |
HS2ST1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr14_-_92040027 | 0.06 |
ENST00000267622.8
|
TRIP11
|
thyroid hormone receptor interactor 11 |
| chr1_-_205631962 | 0.06 |
ENST00000289703.8
ENST00000357992.9 |
ELK4
|
ETS transcription factor ELK4 |
| chr6_+_46652968 | 0.06 |
ENST00000371347.10
|
SLC25A27
|
solute carrier family 25 member 27 |
| chr6_-_159044980 | 0.06 |
ENST00000367066.8
|
TAGAP
|
T cell activation RhoGTPase activating protein |
| chr20_+_35226676 | 0.06 |
ENST00000246186.8
|
MMP24
|
matrix metallopeptidase 24 |
| chr20_+_45890236 | 0.06 |
ENST00000372484.8
ENST00000677394.1 |
CTSA
|
cathepsin A |
| chr1_-_165356703 | 0.06 |
ENST00000342310.7
|
LMX1A
|
LIM homeobox transcription factor 1 alpha |
| chr16_+_1351923 | 0.06 |
ENST00000204679.9
ENST00000527137.2 ENST00000683887.1 ENST00000529110.2 |
GNPTG
|
N-acetylglucosamine-1-phosphate transferase subunit gamma |
| chr6_-_52284677 | 0.06 |
ENST00000596288.7
ENST00000616552.4 ENST00000229854.12 ENST00000419835.8 |
MCM3
|
minichromosome maintenance complex component 3 |
| chr5_+_134371561 | 0.06 |
ENST00000265339.7
ENST00000506787.5 ENST00000507277.1 |
UBE2B
|
ubiquitin conjugating enzyme E2 B |
| chr15_-_61229297 | 0.06 |
ENST00000335670.11
|
RORA
|
RAR related orphan receptor A |
| chr12_-_122526929 | 0.05 |
ENST00000331738.12
ENST00000528279.1 ENST00000344591.8 ENST00000526560.6 |
RSRC2
|
arginine and serine rich coiled-coil 2 |
| chr1_+_35883189 | 0.05 |
ENST00000674304.1
ENST00000373204.6 ENST00000674426.1 |
AGO1
|
argonaute RISC component 1 |
| chr17_+_12665882 | 0.05 |
ENST00000425538.6
|
MYOCD
|
myocardin |
| chr3_-_146251068 | 0.05 |
ENST00000433593.6
ENST00000476202.5 ENST00000354952.7 ENST00000460885.5 |
PLSCR4
|
phospholipid scramblase 4 |
| chr19_+_4402615 | 0.05 |
ENST00000301280.10
|
CHAF1A
|
chromatin assembly factor 1 subunit A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.3 | 1.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.3 | 0.8 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.2 | 0.6 | GO:1903892 | negative regulation of ATF6-mediated unfolded protein response(GO:1903892) |
| 0.2 | 0.8 | GO:0035948 | N-terminal peptidyl-lysine acetylation(GO:0018076) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.3 | GO:0071464 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) cellular response to hydrostatic pressure(GO:0071464) |
| 0.1 | 0.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 1.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.5 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 1.0 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.2 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.2 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.3 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.2 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.4 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.2 | GO:2000979 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.5 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 0.2 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.2 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.1 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.1 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.4 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.2 | GO:0048669 | microglia differentiation(GO:0014004) microglia development(GO:0014005) collateral sprouting in absence of injury(GO:0048669) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 | 0.5 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.3 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0030221 | basophil differentiation(GO:0030221) astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.7 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.1 | GO:0030806 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.0 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.2 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.2 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.4 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.3 | GO:0072498 | cornea development in camera-type eye(GO:0061303) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.2 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.2 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.0 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.8 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 1.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0097135 | X chromosome(GO:0000805) cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.1 | GO:0002081 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.8 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.3 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.2 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 0.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.3 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.5 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.2 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.4 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0038064 | collagen receptor activity(GO:0038064) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.0 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |