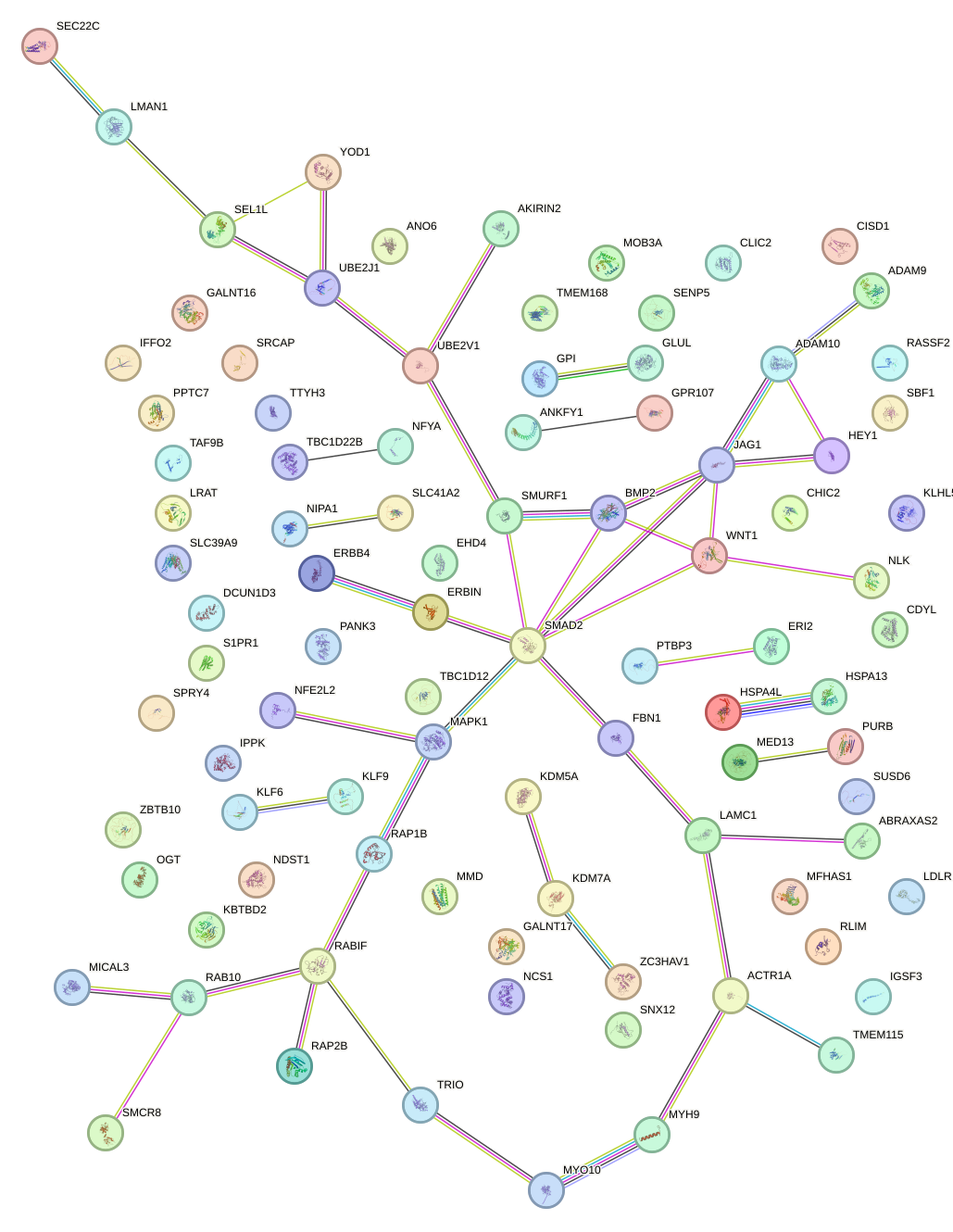
chr5_-_142325001
Show fit
0.85
ENST00000344120.4
ENST00000434127.3
SPRY4
sprouty RTK signaling antagonist 4
chr20_+_6767678
Show fit
0.74
ENST00000378827.5
BMP2
bone morphogenetic protein 2
chr8_+_80485641
Show fit
0.64
ENST00000430430.5
ZBTB10
zinc finger and BTB domain containing 10
chr9_-_70414657
Show fit
0.59
ENST00000377126.4
KLF9
Kruppel like factor 9
chr12_-_110583305
Show fit
0.56
ENST00000354300.5
PPTC7
protein phosphatase targeting COQ7
chr12_-_104958268
Show fit
0.48
ENST00000432951.1
ENST00000258538.8
ENST00000415674.1
ENST00000424946.1
ENST00000433540.5
SLC41A2
solute carrier family 41 member 2
chr12_+_45216079
Show fit
0.44
ENST00000423947.7
ENST00000680498.1
ENST00000320560.13
ANO6
anoctamin 6
chr20_-_10673987
Show fit
0.43
ENST00000254958.10
JAG1
jagged canonical Notch ligand 1
chr8_-_79767462
Show fit
0.41
ENST00000674295.1
ENST00000518733.1
ENST00000674418.1
ENST00000674358.1
ENST00000354724.8
HEY1
hes related family bHLH transcription factor with YRPW motif 1
chr6_+_4889992
Show fit
0.40
ENST00000343762.5
CDYL
chromodomain Y like
chr17_-_55421818
Show fit
0.39
ENST00000262065.8
ENST00000649377.1
MMD
monocyte to macrophage differentiation associated
chr7_-_99144053
Show fit
0.36
ENST00000361125.1
ENST00000361368.7
SMURF1
SMAD specific E3 ubiquitin protein ligase 1
chr14_+_69611586
Show fit
0.36
ENST00000342745.5
SUSD6
sushi domain containing 6
chr7_-_32891744
Show fit
0.35
ENST00000304056.9
KBTBD2
kelch repeat and BTB domain containing 2
chr8_-_8893548
Show fit
0.35
ENST00000276282.7
MFHAS1
malignant fibrous histiocytoma amplified sequence 1
chr6_-_89352706
Show fit
0.29
ENST00000435041.3
UBE2J1
ubiquitin conjugating enzyme E2 J1
chr9_+_99105098
Show fit
0.29
ENST00000374990.6
ENST00000374994.9
ENST00000552516.5
TGFBR1
transforming growth factor beta receptor 1
chr9_+_110048598
Show fit
0.29
ENST00000434623.6
ENST00000374525.5
PALM2AKAP2
PALM2 and AKAP2 fusion
chrX_-_155334580
Show fit
0.28
ENST00000369449.7
ENST00000321926.4
CLIC2
chloride intracellular channel 2
chr21_-_14383125
Show fit
0.27
ENST00000285667.4
HSPA13
heat shock protein family A (Hsp70) member 13
chr17_-_35121487
Show fit
0.26
ENST00000593039.5
ENSG00000267618.5
RAD51L3-RFFL readthrough
chr15_-_41972504
Show fit
0.24
ENST00000220325.9
EHD4
EH domain containing 4
chr9_+_109780292
Show fit
0.23
ENST00000374530.7
PALM2AKAP2
PALM2 and AKAP2 fusion
chr5_+_14143322
Show fit
0.23
ENST00000344204.9
TRIO
trio Rho guanine nucleotide exchange factor
chr9_-_112333603
Show fit
0.22
ENST00000450374.1
ENST00000374257.6
ENST00000374255.6
ENST00000334318.10
PTBP3
polypyrimidine tract binding protein 3
chr1_-_182391783
Show fit
0.22
ENST00000331872.11
ENST00000339526.8
GLUL
glutamate-ammonia ligase
chr22_-_18024513
Show fit
0.21
ENST00000441493.7
MICAL3
microtubule associated monooxygenase, calponin and LIM domain containing 3
chr15_+_22786610
Show fit
0.20
ENST00000337435.9
NIPA1
NIPA magnesium transporter 1
chr20_-_31722533
Show fit
0.20
ENST00000677194.1
ENST00000434194.2
ENST00000376062.6
BCL2L1
BCL2 like 1
chr5_+_150508110
Show fit
0.20
ENST00000261797.7
NDST1
N-deacetylase and N-sulfotransferase 1
chr9_+_130172343
Show fit
0.19
ENST00000372398.6
NCS1
neuronal calcium sensor 1
chr6_+_37257762
Show fit
0.19
ENST00000373491.3
TBC1D22B
TBC1 domain family member 22B
chr10_+_58269132
Show fit
0.19
ENST00000333926.6
CISD1
CDGSH iron sulfur domain 1
chr2_+_241315882
Show fit
0.18
ENST00000401990.5
ENST00000407971.5
ENST00000436795.5
ENST00000411484.5
ENST00000434955.5
ENST00000391971.7
ENST00000402092.6
ENST00000441533.5
ENST00000443492.5
ENST00000437066.5
ENST00000429791.5
SEPTIN2
septin 2
chr2_+_26034069
Show fit
0.18
ENST00000264710.5
RAB10
RAB10, member RAS oncogene family
chrX_+_71533095
Show fit
0.18
ENST00000373719.8
ENST00000373701.7
OGT
O-linked N-acetylglucosamine (GlcNAc) transferase
chr1_+_183023409
Show fit
0.17
ENST00000258341.5
LAMC1
laminin subunit gamma 1
chr6_-_81752671
Show fit
0.17
ENST00000320172.11
ENST00000369754.7
ENST00000369756.3
TENT5A
terminal nucleotidyltransferase 5A
chr2_-_212538766
Show fit
0.16
ENST00000342788.9
ERBB4
erb-b2 receptor tyrosine kinase 4
chr9_-_92670124
Show fit
0.16
ENST00000287996.8
IPPK
inositol-pentakisphosphate 2-kinase
chr22_-_21867610
Show fit
0.16
ENST00000215832.11
ENST00000398822.7
MAPK1
mitogen-activated protein kinase 1
chr6_+_41072939
Show fit
0.16
ENST00000341376.11
ENST00000353205.5
NFYA
nuclear transcription factor Y subunit alpha
chr16_-_20900319
Show fit
0.16
ENST00000564349.5
ENST00000324344.9
ERI2
DCUN1D3
ERI1 exoribonuclease family member 2
chr7_+_2631978
Show fit
0.15
ENST00000258796.12
TTYH3
tweety family member 3
chr3_-_42581936
Show fit
0.15
ENST00000423701.6
ENST00000420163.1
ENST00000416880.5
ENST00000264454.8
ENST00000273156.11
SEC22C
SEC22 homolog C, vesicle trafficking protein
chr20_-_4823597
Show fit
0.15
ENST00000379400.8
RASSF2
Ras association domain family member 2
chr17_+_28042660
Show fit
0.15
ENST00000407008.8
NLK
nemo like kinase
chr22_-_36387949
Show fit
0.15
ENST00000216181.11
MYH9
myosin heavy chain 9
chr1_-_116667668
Show fit
0.15
ENST00000369486.8
ENST00000369483.5
IGSF3
immunoglobulin superfamily member 3
chr4_-_54064586
Show fit
0.14
ENST00000263921.8
CHIC2
cysteine rich hydrophobic domain 2
chr10_-_3785225
Show fit
0.14
ENST00000542957.1
KLF6
Kruppel like factor 6
chr8_+_38996899
Show fit
0.14
ENST00000677582.1
ENST00000676643.1
ENST00000676936.1
ENST00000677004.1
ENST00000487273.7
ENST00000481513.5
ADAM9
ADAM metallopeptidase domain 9
chr6_-_87702221
Show fit
0.14
ENST00000257787.6
AKIRIN2
akirin 2
chr5_-_16936231
Show fit
0.14
ENST00000507288.1
ENST00000274203.13
ENST00000513610.6
MYO10
myosin X
chr22_-_50474942
Show fit
0.13
ENST00000348911.10
ENST00000380817.8
SBF1
SET binding factor 1
chr12_+_48978313
Show fit
0.13
ENST00000293549.4
WNT1
Wnt family member 1
chr12_-_389249
Show fit
0.13
ENST00000535014.5
ENST00000543507.1
ENST00000544760.1
ENST00000399788.7
KDM5A
lysine demethylase 5A
chr5_-_168579319
Show fit
0.13
ENST00000522176.1
ENST00000239231.7
PANK3
pantothenate kinase 3
chr5_+_65926556
Show fit
0.13
ENST00000380943.6
ENST00000416865.6
ENST00000380935.5
ENST00000284037.10
ERBIN
erbb2 interacting protein
chr10_-_102502669
Show fit
0.13
ENST00000487599.1
ACTR1A
actin related protein 1A
chr4_-_110636963
Show fit
0.13
ENST00000394595.8
PITX2
paired like homeodomain 2
chr18_-_59359245
Show fit
0.13
ENST00000251047.6
LMAN1
lectin, mannose binding 1
chr3_-_52679713
Show fit
0.12
ENST00000296302.11
ENST00000356770.8
ENST00000337303.8
ENST00000409057.5
ENST00000410007.5
ENST00000409114.7
ENST00000409767.5
ENST00000423351.5
PBRM1
polybromo 1
chr17_-_62065248
Show fit
0.12
ENST00000397786.7
MED13
mediator complex subunit 13
chr3_+_196867856
Show fit
0.12
ENST00000445299.6
ENST00000323460.10
ENST00000419026.5
SENP5
SUMO specific peptidase 5
chr10_+_124801799
Show fit
0.12
ENST00000298492.6
ABRAXAS2
abraxas 2, BRISC complex subunit
chr3_+_153162196
Show fit
0.12
ENST00000323534.5
RAP2B
RAP2B, member of RAS oncogene family
chr19_+_11089446
Show fit
0.11
ENST00000557933.5
ENST00000455727.6
ENST00000535915.5
ENST00000545707.5
ENST00000558518.6
ENST00000558013.5
LDLR
low density lipoprotein receptor
chr14_-_81533800
Show fit
0.11
ENST00000555824.5
ENST00000557372.1
ENST00000336735.9
SEL1L
SEL1L adaptor subunit of ERAD E3 ubiquitin ligase
chr2_-_177263800
Show fit
0.11
ENST00000397063.8
ENST00000421929.5
NFE2L2
nuclear factor, erythroid 2 like 2
chr18_-_57586668
Show fit
0.11
ENST00000592699.6
ENST00000382873.8
ENST00000262093.11
ENST00000652755.1
FECH
ferrochelatase
chr15_-_58749569
Show fit
0.10
ENST00000402627.5
ENST00000559053.1
ENST00000260408.8
ENST00000561288.1
ENST00000461408.2
ENST00000439637.5
ENST00000558004.1
ADAM10
ADAM metallopeptidase domain 10
chr17_+_18315273
Show fit
0.10
ENST00000406438.5
SMCR8
SMCR8-C9orf72 complex subunit
chr7_-_140176970
Show fit
0.10
ENST00000397560.7
KDM7A
lysine demethylase 7A
chr3_-_50359480
Show fit
0.10
ENST00000266025.4
TMEM115
transmembrane protein 115
chr10_+_94402486
Show fit
0.10
ENST00000225235.5
TBC1D12
TBC1 domain family member 12
chr1_-_18956669
Show fit
0.10
ENST00000455833.7
IFFO2
intermediate filament family orphan 2
chr17_-_4263847
Show fit
0.09
ENST00000570535.5
ENST00000574367.5
ENST00000341657.9
ANKFY1
ankyrin repeat and FYVE domain containing 1
chr4_+_127782270
Show fit
0.09
ENST00000508549.5
ENST00000296464.9
HSPA4L
heat shock protein family A (Hsp70) member 4 like
chr6_+_43770707
Show fit
0.09
ENST00000324450.11
ENST00000417285.7
ENST00000413642.8
ENST00000372055.9
ENST00000482630.7
ENST00000425836.7
ENST00000372064.9
ENST00000372077.8
ENST00000519767.5
VEGFA
vascular endothelial growth factor A
chr1_+_101237009
Show fit
0.09
ENST00000305352.7
S1PR1
sphingosine-1-phosphate receptor 1
chr1_-_202889099
Show fit
0.09
ENST00000367262.4
RABIF
RAB interacting factor
chr12_+_68610858
Show fit
0.09
ENST00000393436.9
ENST00000425247.6
ENST00000250559.14
ENST00000489473.6
ENST00000422358.6
ENST00000541167.5
ENST00000538283.5
ENST00000341355.9
ENST00000537460.5
ENST00000450214.6
ENST00000545270.5
RAP1B
RAP1B, member of RAS oncogene family
chr7_-_139109702
Show fit
0.09
ENST00000471652.1
ENST00000242351.10
ZC3HAV1
zinc finger CCCH-type containing, antiviral 1
chr4_+_154743993
Show fit
0.09
ENST00000336356.4
LRAT
lecithin retinol acyltransferase
chr16_+_30699155
Show fit
0.09
ENST00000262518.9
SRCAP
Snf2 related CREBBP activator protein
chr15_-_48645701
Show fit
0.09
ENST00000316623.10
ENST00000560355.1
FBN1
fibrillin 1
chr4_-_145938422
Show fit
0.09
ENST00000656985.1
ENST00000652097.1
ENST00000503462.3
ENST00000379448.9
ENST00000513840.2
ZNF827
zinc finger protein 827
chr1_-_207051202
Show fit
0.08
ENST00000315927.9
YOD1
YOD1 deubiquitinase
chr7_-_112790372
Show fit
0.08
ENST00000449743.1
ENST00000441474.1
ENST00000312814.11
ENST00000454074.5
ENST00000447395.5
TMEM168
transmembrane protein 168
chrX_-_71068311
Show fit
0.08
ENST00000374274.8
SNX12
sorting nexin 12
chr7_-_44885446
Show fit
0.08
ENST00000395699.5
PURB
purine rich element binding protein B
chrX_-_78139612
Show fit
0.08
ENST00000341864.6
TAF9B
TATA-box binding protein associated factor 9b
chr4_+_39044995
Show fit
0.08
ENST00000261425.7
ENST00000508137.6
KLHL5
kelch like family member 5
chr18_-_47930630
Show fit
0.07
ENST00000262160.11
SMAD2
SMAD family member 2
chr2_+_207711631
Show fit
0.07
ENST00000295414.8
ENST00000420822.1
ENST00000339882.9
CCNYL1
cyclin Y like 1
chr19_-_2096260
Show fit
0.07
ENST00000588048.2
ENST00000357066.8
ENST00000591236.1
MOB3A
MOB kinase activator 3A
chr11_-_31811314
Show fit
0.07
ENST00000640368.2
ENST00000379123.10
ENST00000379115.9
ENST00000419022.6
ENST00000643871.1
ENST00000640610.1
ENST00000639034.2
PAX6
paired box 6
chrX_-_74614612
Show fit
0.07
ENST00000349225.2
ENST00000332687.11
RLIM
ring finger protein, LIM domain interacting
chr9_+_130053706
Show fit
0.06
ENST00000372410.7
GPR107
G protein-coupled receptor 107
chr14_+_69259937
Show fit
0.06
ENST00000337827.8
GALNT16
polypeptide N-acetylgalactosaminyltransferase 16
chr20_-_50113139
Show fit
0.06
ENST00000371657.9
ENST00000371674.8
ENST00000625172.3
ENST00000557021.5
ENST00000617119.4
UBE2V1
ubiquitin conjugating enzyme E2 V1
chr14_+_69398683
Show fit
0.06
ENST00000556605.5
ENST00000031146.8
ENST00000336643.10
SLC39A9
solute carrier family 39 member 9
chr22_+_19131271
Show fit
0.06
ENST00000399635.4
TSSK2
testis specific serine kinase 2
chr1_-_67833448
Show fit
0.06
ENST00000370982.4
GNG12
G protein subunit gamma 12
chr17_-_35089212
Show fit
0.06
ENST00000584655.5
ENST00000447669.6
ENST00000315249.11
RFFL
ring finger and FYVE like domain containing E3 ubiquitin protein ligase
chr1_+_25338294
Show fit
0.06
ENST00000374358.5
TMEM50A
transmembrane protein 50A
chr4_+_183905266
Show fit
0.06
ENST00000308497.9
STOX2
storkhead box 2
chr3_+_15427551
Show fit
0.06
ENST00000396842.7
EAF1
ELL associated factor 1
chr5_-_56952107
Show fit
0.06
ENST00000381226.7
ENST00000381199.8
ENST00000381213.7
MIER3
MIER family member 3
chr2_+_114442616
Show fit
0.06
ENST00000410059.6
DPP10
dipeptidyl peptidase like 10
chr5_-_34915493
Show fit
0.06
ENST00000382038.7
RAD1
RAD1 checkpoint DNA exonuclease
chr19_+_39125769
Show fit
0.05
ENST00000602004.1
ENST00000599470.5
ENST00000321944.8
ENST00000593480.5
ENST00000358301.7
ENST00000593690.5
ENST00000599386.5
PAK4
p21 (RAC1) activated kinase 4
chr12_-_96400365
Show fit
0.05
ENST00000261211.8
ENST00000543119.6
CDK17
cyclin dependent kinase 17
chr14_+_23306816
Show fit
0.05
ENST00000678311.1
ENST00000557579.2
ENST00000250405.10
ENST00000679000.1
ENST00000557236.6
ENST00000678502.1
ENST00000553781.5
BCL2L2
BCL2L2-PABPN1
BCL2 like 2
chr4_+_54229261
Show fit
0.05
ENST00000508170.5
ENST00000512143.1
ENST00000257290.10
PDGFRA
platelet derived growth factor receptor alpha
chr7_+_39623547
Show fit
0.05
ENST00000005257.7
RALA
RAS like proto-oncogene A
chr12_+_130872037
Show fit
0.05
ENST00000448750.7
ENST00000541630.5
ENST00000543796.6
ENST00000392369.6
ENST00000535090.5
ENST00000541679.7
ENST00000392367.4
RAN
RAN, member RAS oncogene family
chr1_-_174022339
Show fit
0.04
ENST00000367696.7
RC3H1
ring finger and CCCH-type domains 1
chr12_-_56258327
Show fit
0.04
ENST00000267116.8
ANKRD52
ankyrin repeat domain 52
chr21_+_29298890
Show fit
0.04
ENST00000286800.8
BACH1
BTB domain and CNC homolog 1
chr15_-_83207800
Show fit
0.04
ENST00000299633.7
HDGFL3
HDGF like 3
chr18_+_34493289
Show fit
0.04
ENST00000682923.1
ENST00000596745.5
ENST00000283365.14
ENST00000315456.10
ENST00000598774.6
ENST00000684266.1
ENST00000683092.1
ENST00000683379.1
ENST00000684359.1
DTNA
dystrobrevin alpha
chr19_-_40690629
Show fit
0.04
ENST00000252891.8
NUMBL
NUMB like endocytic adaptor protein
chr4_-_151226427
Show fit
0.04
ENST00000304527.8
ENST00000409598.8
SH3D19
SH3 domain containing 19
chr7_+_128739292
Show fit
0.04
ENST00000535011.6
ENST00000542996.6
ENST00000249364.9
ENST00000449187.6
CALU
calumenin
chr3_+_57756230
Show fit
0.04
ENST00000295951.7
ENST00000659705.1
ENST00000671191.1
SLMAP
sarcolemma associated protein
chr1_+_171841466
Show fit
0.03
ENST00000367733.6
ENST00000627582.3
ENST00000355305.9
ENST00000367731.5
DNM3
dynamin 3
chr7_-_117873420
Show fit
0.03
ENST00000160373.8
CTTNBP2
cortactin binding protein 2
chr7_+_92245960
Show fit
0.03
ENST00000265742.8
ANKIB1
ankyrin repeat and IBR domain containing 1
chr7_-_113087720
Show fit
0.03
ENST00000297146.7
GPR85
G protein-coupled receptor 85
chr20_+_45363114
Show fit
0.03
ENST00000426004.5
ENST00000243918.10
SYS1
SYS1 golgi trafficking protein
chr2_-_224585354
Show fit
0.03
ENST00000264414.9
ENST00000344951.8
CUL3
cullin 3
chrX_+_37571569
Show fit
0.03
ENST00000614025.4
ENST00000378621.7
ENST00000378619.4
LANCL3
LanC like 3
chr7_-_76358982
Show fit
0.03
ENST00000307630.5
YWHAG
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma
chr16_-_66751591
Show fit
0.03
ENST00000440564.6
ENST00000443351.6
ENST00000566150.5
ENST00000258198.7
DYNC1LI2
dynein cytoplasmic 1 light intermediate chain 2
chr6_-_31660735
Show fit
0.03
ENST00000375911.2
C6orf47
chromosome 6 open reading frame 47
chr21_-_26967057
Show fit
0.03
ENST00000284987.6
ADAMTS5
ADAM metallopeptidase with thrombospondin type 1 motif 5
chr20_+_50731571
Show fit
0.03
ENST00000371610.7
PARD6B
par-6 family cell polarity regulator beta
chr16_+_69187125
Show fit
0.03
ENST00000336278.8
SNTB2
syntrophin beta 2
chr14_+_64704380
Show fit
0.03
ENST00000247226.13
ENST00000394691.7
PLEKHG3
pleckstrin homology and RhoGEF domain containing G3
chr10_+_102776237
Show fit
0.03
ENST00000369889.5
WBP1L
WW domain binding protein 1 like
chr11_-_46918522
Show fit
0.03
ENST00000378623.6
ENST00000534404.1
LRP4
LDL receptor related protein 4
chr20_+_20017303
Show fit
0.02
ENST00000310450.8
ENST00000334982.9
ENST00000398602.2
NAA20
N-alpha-acetyltransferase 20, NatB catalytic subunit
chr19_-_4066892
Show fit
0.02
ENST00000322357.9
ZBTB7A
zinc finger and BTB domain containing 7A
chr10_+_96832252
Show fit
0.02
ENST00000676187.1
ENST00000675687.1
ENST00000676123.1
ENST00000675471.1
ENST00000371103.8
ENST00000421806.4
ENST00000675250.1
ENST00000540664.6
ENST00000676414.1
LCOR
ligand dependent nuclear receptor corepressor
chr15_+_90001300
Show fit
0.02
ENST00000268154.9
ZNF710
zinc finger protein 710
chr9_-_97039102
Show fit
0.02
ENST00000538255.6
ENST00000680221.1
ENST00000681737.1
ENST00000259470.6
ENST00000681927.1
CTSV
cathepsin V
chr4_-_173530219
Show fit
0.02
ENST00000359562.4
HAND2
Links
heart and neural crest derivatives expressed 2
chr1_+_160343375
Show fit
0.02
ENST00000294785.10
ENST00000421914.5
ENST00000438008.5
NCSTN
nicastrin
chr18_+_9136757
Show fit
0.02
ENST00000262126.9
ENST00000577992.1
ANKRD12
ankyrin repeat domain 12
chr11_+_61792878
Show fit
0.02
ENST00000305885.3
ENST00000535723.1
FEN1
flap structure-specific endonuclease 1
chrX_+_37349287
Show fit
0.02
ENST00000466533.5
ENST00000542554.5
ENST00000543642.5
ENST00000484460.5
ENST00000378628.9
ENST00000449135.6
ENST00000463135.1
ENST00000465127.1
PRRG1
ENSG00000250349.3
proline rich and Gla domain 1
chr2_+_207529892
Show fit
0.02
ENST00000432329.6
ENST00000445803.5
CREB1
cAMP responsive element binding protein 1
chr2_+_53787030
Show fit
0.02
ENST00000185150.9
ENST00000405123.7
ENST00000378239.5
ERLEC1
endoplasmic reticulum lectin 1
chr15_-_29822418
Show fit
0.02
ENST00000614355.5
ENST00000495972.6
ENST00000346128.10
TJP1
tight junction protein 1
chr4_+_17614630
Show fit
0.01
ENST00000237380.12
MED28
mediator complex subunit 28
chr20_+_1118590
Show fit
0.01
ENST00000246015.8
ENST00000335877.11
PSMF1
proteasome inhibitor subunit 1
chr13_+_42048645
Show fit
0.01
ENST00000337343.9
ENST00000261491.9
ENST00000611224.1
DGKH
diacylglycerol kinase eta
chr5_-_90529511
Show fit
0.01
ENST00000500869.6
ENST00000315948.11
ENST00000509384.5
LYSMD3
LysM domain containing 3
chr9_-_23821275
Show fit
0.01
ENST00000380110.8
ELAVL2
ELAV like RNA binding protein 2
chr8_-_66667138
Show fit
0.01
ENST00000310421.5
VCPIP1
valosin containing protein interacting protein 1
chr20_+_33662310
Show fit
0.01
ENST00000375222.4
C20orf144
chromosome 20 open reading frame 144
chr14_-_74019255
Show fit
0.01
ENST00000334696.11
ENST00000556242.5
ENTPD5
ectonucleoside triphosphate diphosphohydrolase 5 (inactive)
chr7_+_114414997
Show fit
0.01
ENST00000462331.5
ENST00000393491.7
ENST00000403559.8
ENST00000408937.7
ENST00000393498.6
ENST00000393495.7
ENST00000378237.7
FOXP2
forkhead box P2
chrX_-_84188148
Show fit
0.01
ENST00000262752.5
RPS6KA6
ribosomal protein S6 kinase A6
chr15_+_80779343
Show fit
0.01
ENST00000220244.7
ENST00000394685.8
ENST00000356249.9
CEMIP
cell migration inducing hyaluronidase 1
chr1_+_26021768
Show fit
0.01
ENST00000374280.4
EXTL1
exostosin like glycosyltransferase 1
chr20_-_35742207
Show fit
0.01
ENST00000397370.3
ENST00000528062.7
ENST00000374038.7
ENST00000253363.11
ENST00000361162.10
RBM39
RNA binding motif protein 39
chr1_-_42958836
Show fit
0.01
ENST00000372500.4
ENST00000674765.1
ENST00000460369.3
ENST00000426263.10
SLC2A1
solute carrier family 2 member 1
chr2_-_199457931
Show fit
0.01
ENST00000417098.6
SATB2
SATB homeobox 2
chr8_+_69466617
Show fit
0.01
ENST00000525061.5
ENST00000260128.8
ENST00000458141.6
SULF1
sulfatase 1
chr5_+_32711313
Show fit
0.01
ENST00000265074.13
NPR3
natriuretic peptide receptor 3
chr9_-_104928139
Show fit
0.01
ENST00000423487.6
ENST00000374733.1
ENST00000374736.8
ENST00000678995.1
ABCA1
ATP binding cassette subfamily A member 1
chr19_+_9835292
Show fit
0.00
ENST00000247970.9
ENST00000587625.5
ENST00000588695.5
PIN1
peptidylprolyl cis/trans isomerase, NIMA-interacting 1
chr12_-_48788995
Show fit
0.00
ENST00000550422.5
ENST00000357869.8
ADCY6
adenylate cyclase 6
chr22_-_24555086
Show fit
0.00
ENST00000447813.6
ENST00000402766.5
ENST00000435822.6
ENST00000407471.7
GUCD1
guanylyl cyclase domain containing 1
chr9_+_5629025
Show fit
0.00
ENST00000251879.10
ENST00000414202.7
ENST00000418622.7
RIC1
RIC1 homolog, RAB6A GEF complex partner 1
chr20_-_50153637
Show fit
0.00
ENST00000341698.2
ENST00000371652.9
ENST00000371650.9
PEDS1-UBE2V1
PEDS1
PEDS1-UBE2V1 readthrough
chr2_-_37156942
Show fit
0.00
ENST00000680273.1
ENST00000233057.9
ENST00000679979.1
ENST00000679507.1
ENST00000681463.1
ENST00000395127.6
EIF2AK2
eukaryotic translation initiation factor 2 alpha kinase 2
chr6_+_148747016
Show fit
0.00
ENST00000367463.5
UST
uronyl 2-sulfotransferase
chr18_-_26865689
Show fit
0.00
ENST00000675739.1
ENST00000383168.9
ENST00000672981.2
ENST00000578776.1
AQP4
aquaporin 4
chr13_-_25172278
Show fit
0.00
ENST00000515384.2
ENST00000357816.2
AMER2
APC membrane recruitment protein 2
chr4_-_23890035
Show fit
0.00
ENST00000507380.1
ENST00000264867.7
PPARGC1A
PPARG coactivator 1 alpha