Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
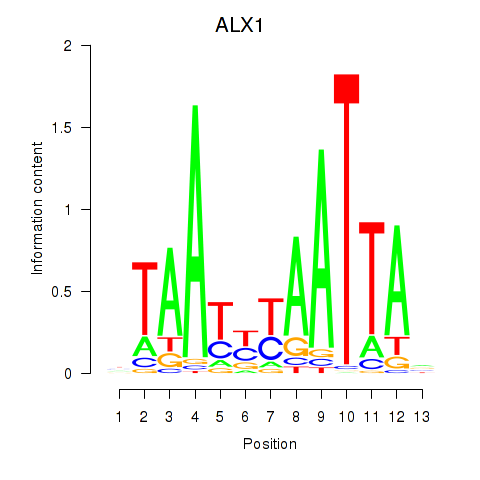
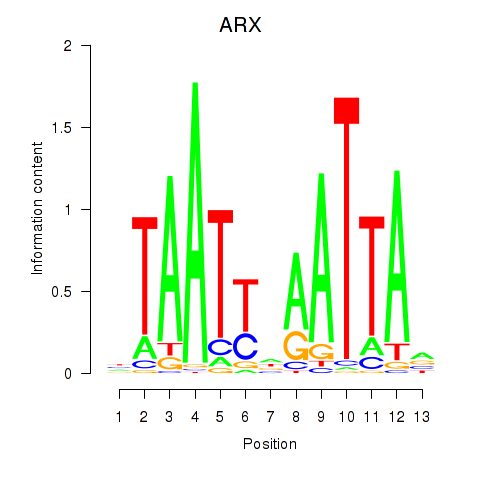
Results for ALX1_ARX
Z-value: 0.30
Transcription factors associated with ALX1_ARX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX1
|
ENSG00000180318.4 | ALX1 |
|
ARX
|
ENSG00000004848.8 | ARX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARX | hg38_v1_chrX_-_25015924_25016002 | -0.43 | 3.3e-02 | Click! |
| ALX1 | hg38_v1_chr12_+_85280199_85280237 | 0.23 | 2.7e-01 | Click! |
Activity profile of ALX1_ARX motif
Sorted Z-values of ALX1_ARX motif
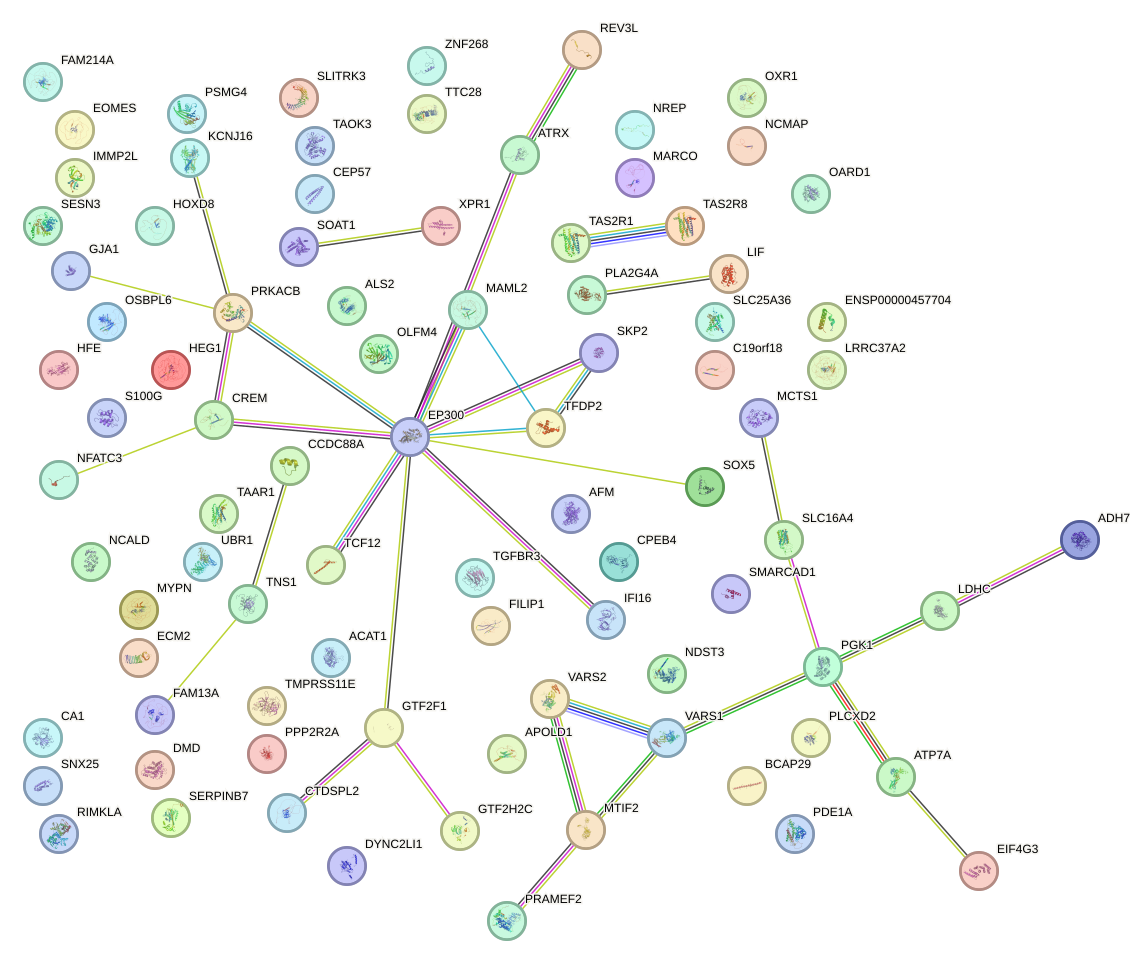
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX1_ARX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_55269038 | 0.91 |
ENST00000417363.5
ENST00000412530.1 ENST00000366137.6 ENST00000420637.5 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr1_-_91906280 | 0.54 |
ENST00000370399.6
|
TGFBR3
|
transforming growth factor beta receptor 3 |
| chr6_+_121437378 | 0.36 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr6_-_132659178 | 0.34 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr3_-_165196689 | 0.30 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr6_-_41071825 | 0.29 |
ENST00000468811.5
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr1_+_84164962 | 0.29 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr3_-_165196369 | 0.27 |
ENST00000475390.2
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr12_-_118359639 | 0.26 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr1_+_84181630 | 0.25 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr3_-_27722699 | 0.24 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr10_+_68106109 | 0.23 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr15_+_48191648 | 0.21 |
ENST00000646012.1
ENST00000561127.5 ENST00000647546.1 ENST00000559641.5 ENST00000417307.3 |
SLC12A1
CTXN2
|
solute carrier family 12 member 1 cortexin 2 |
| chr2_-_55334529 | 0.21 |
ENST00000645860.1
ENST00000642563.1 ENST00000647396.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr3_-_142029108 | 0.21 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr9_+_72577939 | 0.19 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr3_-_33718207 | 0.19 |
ENST00000359576.9
ENST00000682230.1 ENST00000399362.8 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr13_+_53028806 | 0.19 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr6_-_75363003 | 0.18 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr4_-_99435134 | 0.17 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr2_+_43774033 | 0.17 |
ENST00000260605.12
ENST00000406852.7 ENST00000398823.6 ENST00000605786.5 |
DYNC2LI1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr7_-_111392915 | 0.16 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr3_-_125055987 | 0.16 |
ENST00000311127.9
|
HEG1
|
heart development protein with EGF like domains 1 |
| chr3_-_27722316 | 0.16 |
ENST00000449599.4
|
EOMES
|
eomesodermin |
| chr1_+_159008978 | 0.15 |
ENST00000447473.6
|
IFI16
|
interferon gamma inducible protein 16 |
| chr2_+_176129680 | 0.15 |
ENST00000429017.2
ENST00000313173.6 |
HOXD8
|
homeobox D8 |
| chrX_+_77910656 | 0.15 |
ENST00000343533.9
ENST00000341514.11 ENST00000645454.1 ENST00000642651.1 ENST00000644362.1 |
ATP7A
PGK1
|
ATPase copper transporting alpha phosphoglycerate kinase 1 |
| chr9_+_72577369 | 0.14 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr4_+_185204237 | 0.14 |
ENST00000618785.4
ENST00000504273.5 |
SNX25
|
sorting nexin 25 |
| chr6_-_111483700 | 0.14 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr16_+_68085344 | 0.14 |
ENST00000575270.5
ENST00000346183.8 |
NFATC3
|
nuclear factor of activated T cells 3 |
| chr15_+_44427793 | 0.13 |
ENST00000558966.5
ENST00000559793.5 ENST00000558968.1 |
CTDSPL2
|
CTD small phosphatase like 2 |
| chr1_+_24556087 | 0.13 |
ENST00000374392.3
|
NCMAP
|
non-compact myelin associated protein |
| chr11_-_96343170 | 0.13 |
ENST00000524717.6
|
MAML2
|
mastermind like transcriptional coactivator 2 |
| chr16_+_68085420 | 0.13 |
ENST00000349223.9
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr11_+_95789965 | 0.12 |
ENST00000537677.5
|
CEP57
|
centrosomal protein 57 |
| chr9_+_72577788 | 0.12 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr1_+_248508073 | 0.12 |
ENST00000641804.1
|
OR2G6
|
olfactory receptor family 2 subfamily G member 6 |
| chr5_-_111976925 | 0.12 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr11_+_18412292 | 0.11 |
ENST00000541669.6
ENST00000280704.8 |
LDHC
|
lactate dehydrogenase C |
| chr8_+_106726012 | 0.11 |
ENST00000449762.6
ENST00000297447.10 |
OXR1
|
oxidation resistance 1 |
| chr12_+_59689337 | 0.11 |
ENST00000261187.8
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr15_+_57219411 | 0.10 |
ENST00000543579.5
ENST00000537840.5 ENST00000343827.7 |
TCF12
|
transcription factor 12 |
| chr16_+_68085552 | 0.10 |
ENST00000329524.8
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr9_-_123184233 | 0.10 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr2_-_182427014 | 0.10 |
ENST00000409365.5
ENST00000351439.9 |
PDE1A
|
phosphodiesterase 1A |
| chr5_+_69565122 | 0.10 |
ENST00000507595.1
|
GTF2H2C
|
GTF2H2 family member C |
| chr8_-_85341659 | 0.10 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr17_+_46511511 | 0.09 |
ENST00000576629.5
|
LRRC37A2
|
leucine rich repeat containing 37 member A2 |
| chr7_+_107583919 | 0.09 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr4_+_68447453 | 0.09 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr11_+_95790459 | 0.09 |
ENST00000325486.9
ENST00000325542.10 ENST00000544522.5 ENST00000541365.5 |
CEP57
|
centrosomal protein 57 |
| chr1_-_21050952 | 0.09 |
ENST00000264211.12
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma 3 |
| chr12_-_10807286 | 0.09 |
ENST00000240615.3
|
TAS2R8
|
taste 2 receptor member 8 |
| chr11_-_95232514 | 0.09 |
ENST00000634898.1
ENST00000542176.1 ENST00000278499.6 |
SESN3
|
sestrin 3 |
| chr6_+_26087417 | 0.09 |
ENST00000357618.10
ENST00000309234.10 |
HFE
|
homeostatic iron regulator |
| chr12_+_12725897 | 0.09 |
ENST00000326765.10
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr4_-_89029881 | 0.09 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13 member A |
| chr5_+_36152077 | 0.09 |
ENST00000546211.6
ENST00000620197.5 ENST00000678270.1 ENST00000679015.1 ENST00000678580.1 ENST00000274255.11 ENST00000508514.5 |
SKP2
|
S-phase kinase associated protein 2 |
| chrX_-_32412220 | 0.09 |
ENST00000619831.5
|
DMD
|
dystrophin |
| chr1_+_186828941 | 0.09 |
ENST00000367466.4
|
PLA2G4A
|
phospholipase A2 group IVA |
| chrX_+_16650155 | 0.09 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
| chr3_+_140941901 | 0.09 |
ENST00000453248.6
|
SLC25A36
|
solute carrier family 25 member 36 |
| chr12_-_118190510 | 0.09 |
ENST00000540561.5
ENST00000537952.1 ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr18_+_63775369 | 0.08 |
ENST00000540675.5
|
SERPINB7
|
serpin family B member 7 |
| chr19_-_57974527 | 0.08 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr15_-_52678560 | 0.08 |
ENST00000562351.2
ENST00000261844.11 ENST00000399202.8 ENST00000562135.5 |
FAM214A
|
family with sequence similarity 214 member A |
| chr17_+_70075215 | 0.08 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr4_+_118034480 | 0.08 |
ENST00000296499.6
|
NDST3
|
N-deacetylase and N-sulfotransferase 3 |
| chr3_-_142028597 | 0.08 |
ENST00000467667.5
|
TFDP2
|
transcription factor Dp-2 |
| chr22_-_30246739 | 0.08 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr2_+_118942188 | 0.07 |
ENST00000327097.5
|
MARCO
|
macrophage receptor with collagenous structure |
| chr1_+_42380772 | 0.07 |
ENST00000431473.4
ENST00000410070.6 |
RIMKLA
|
ribosomal modification protein rimK like family member A |
| chr6_-_132734692 | 0.07 |
ENST00000509351.5
ENST00000417437.6 ENST00000423615.6 ENST00000427187.6 ENST00000414302.7 ENST00000367927.9 ENST00000450865.2 |
VNN3
|
vanin 3 |
| chr4_+_73481737 | 0.07 |
ENST00000226355.5
|
AFM
|
afamin |
| chr17_+_70075317 | 0.07 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr2_-_201739205 | 0.07 |
ENST00000681558.1
ENST00000681495.1 |
ALS2
|
alsin Rho guanine nucleotide exchange factor ALS2 |
| chr5_+_36151989 | 0.07 |
ENST00000274254.9
|
SKP2
|
S-phase kinase associated protein 2 |
| chr2_+_178320228 | 0.07 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr8_-_101790934 | 0.07 |
ENST00000523645.5
ENST00000520346.1 ENST00000220931.11 ENST00000522448.5 ENST00000522951.5 ENST00000522252.5 ENST00000519098.5 |
NCALD
|
neurocalcin delta |
| chr17_-_62808299 | 0.07 |
ENST00000311269.10
|
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chr3_+_140941792 | 0.06 |
ENST00000446041.6
ENST00000324194.12 ENST00000507429.5 |
SLC25A36
|
solute carrier family 25 member 36 |
| chr17_-_62808339 | 0.06 |
ENST00000583600.5
|
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chr1_-_110391041 | 0.06 |
ENST00000369781.8
ENST00000437429.6 ENST00000541986.5 |
SLC16A4
|
solute carrier family 16 member 4 |
| chrX_-_77634229 | 0.06 |
ENST00000675732.1
|
ATRX
|
ATRX chromatin remodeler |
| chr19_-_6393205 | 0.06 |
ENST00000595047.5
|
GTF2F1
|
general transcription factor IIF subunit 1 |
| chr1_+_180632001 | 0.06 |
ENST00000367590.9
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr1_-_93681829 | 0.06 |
ENST00000260502.11
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr10_+_35175586 | 0.06 |
ENST00000494479.5
ENST00000463314.5 ENST00000342105.7 ENST00000495301.1 |
CREM
|
cAMP responsive element modulator |
| chrX_+_120604084 | 0.06 |
ENST00000371317.10
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr2_-_218010202 | 0.06 |
ENST00000646520.1
|
TNS1
|
tensin 1 |
| chr8_+_26293112 | 0.06 |
ENST00000523925.5
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr16_+_53099100 | 0.06 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr5_-_9630351 | 0.05 |
ENST00000382492.4
|
TAS2R1
|
taste 2 receptor member 1 |
| chrX_+_120604199 | 0.05 |
ENST00000371315.3
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr4_+_94207596 | 0.05 |
ENST00000359052.8
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr20_+_31547367 | 0.05 |
ENST00000394552.3
|
MCTS2P
|
MCTS family member 2, pseudogene |
| chr22_-_28306645 | 0.05 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr12_+_133181529 | 0.05 |
ENST00000541009.6
ENST00000592241.5 |
ZNF268
|
zinc finger protein 268 |
| chr9_-_92536031 | 0.05 |
ENST00000344604.9
ENST00000375540.5 |
ECM2
|
extracellular matrix protein 2 |
| chr11_+_108116688 | 0.05 |
ENST00000672284.1
|
ACAT1
|
acetyl-CoA acetyltransferase 1 |
| chr3_-_33645433 | 0.05 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr8_+_106726115 | 0.05 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1 |
| chr12_+_133181409 | 0.05 |
ENST00000416488.5
ENST00000228289.9 ENST00000541211.6 ENST00000536435.7 ENST00000500625.7 ENST00000539248.6 ENST00000542711.6 ENST00000536899.6 ENST00000542986.6 ENST00000611984.4 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr15_-_43106022 | 0.05 |
ENST00000627960.1
ENST00000290650.9 |
UBR1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr6_+_3258888 | 0.05 |
ENST00000380305.4
|
PSMG4
|
proteasome assembly chaperone 4 |
| chr3_+_111674654 | 0.05 |
ENST00000636933.1
ENST00000393934.7 ENST00000477665.2 |
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2 |
| chr3_-_142028617 | 0.05 |
ENST00000477292.5
ENST00000478006.5 ENST00000495310.5 ENST00000486111.5 |
TFDP2
|
transcription factor Dp-2 |
| chr13_+_48256214 | 0.05 |
ENST00000650237.1
|
ITM2B
|
integral membrane protein 2B |
| chr4_+_94207845 | 0.05 |
ENST00000457823.6
ENST00000354268.9 |
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr9_+_128566741 | 0.04 |
ENST00000630866.1
|
SPTAN1
|
spectrin alpha, non-erythrocytic 1 |
| chr5_+_173918216 | 0.04 |
ENST00000519467.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr4_-_142305935 | 0.04 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr4_+_75724569 | 0.04 |
ENST00000514213.7
ENST00000264904.8 |
USO1
|
USO1 vesicle transport factor |
| chr1_+_12857086 | 0.04 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr22_+_41092869 | 0.04 |
ENST00000674155.1
|
EP300
|
E1A binding protein p300 |
| chr12_-_23584600 | 0.04 |
ENST00000396007.6
|
SOX5
|
SRY-box transcription factor 5 |
| chr6_+_30914205 | 0.04 |
ENST00000672801.1
ENST00000321897.9 ENST00000625423.2 ENST00000676266.1 ENST00000428017.5 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr19_-_7021431 | 0.04 |
ENST00000636986.2
ENST00000637800.1 |
MBD3L2B
|
methyl-CpG binding domain protein 3 like 2B |
| chr2_-_229921316 | 0.04 |
ENST00000428959.5
ENST00000675423.1 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr5_-_56116946 | 0.04 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr1_-_248277976 | 0.04 |
ENST00000641220.1
|
OR2T33
|
olfactory receptor family 2 subfamily T member 33 |
| chr12_-_11134644 | 0.04 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chrX_-_11265975 | 0.04 |
ENST00000303025.10
ENST00000657361.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr2_+_75646775 | 0.04 |
ENST00000393909.7
ENST00000358788.10 ENST00000409374.5 |
MRPL19
|
mitochondrial ribosomal protein L19 |
| chr4_+_94455245 | 0.04 |
ENST00000508216.5
ENST00000514743.5 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr1_+_244051275 | 0.04 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr2_+_67397297 | 0.04 |
ENST00000644028.1
ENST00000272342.6 |
ETAA1
|
ETAA1 activator of ATR kinase |
| chr6_+_30914329 | 0.04 |
ENST00000541562.6
|
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr19_+_55769118 | 0.04 |
ENST00000341750.5
|
RFPL4AL1
|
ret finger protein like 4A like 1 |
| chr3_+_172754457 | 0.04 |
ENST00000441497.6
|
ECT2
|
epithelial cell transforming 2 |
| chr5_-_135399863 | 0.04 |
ENST00000510038.1
ENST00000304332.8 |
MACROH2A1
|
macroH2A.1 histone |
| chrY_-_6874027 | 0.03 |
ENST00000215479.10
|
AMELY
|
amelogenin Y-linked |
| chr15_+_63529142 | 0.03 |
ENST00000268049.11
|
USP3
|
ubiquitin specific peptidase 3 |
| chr14_-_94770102 | 0.03 |
ENST00000238558.5
|
GSC
|
goosecoid homeobox |
| chr3_-_71360753 | 0.03 |
ENST00000648783.1
|
FOXP1
|
forkhead box P1 |
| chr3_-_146528750 | 0.03 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr20_+_33235987 | 0.03 |
ENST00000375422.6
ENST00000375413.8 ENST00000354297.9 |
BPIFA1
|
BPI fold containing family A member 1 |
| chr17_+_76467597 | 0.03 |
ENST00000392492.8
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr11_-_11353241 | 0.03 |
ENST00000528848.3
|
CSNK2A3
|
casein kinase 2 alpha 3 |
| chr11_-_124320197 | 0.03 |
ENST00000624618.2
|
OR8D2
|
olfactory receptor family 8 subfamily D member 2 |
| chr18_-_24311495 | 0.03 |
ENST00000357041.8
|
OSBPL1A
|
oxysterol binding protein like 1A |
| chr2_+_161136901 | 0.03 |
ENST00000259075.6
ENST00000432002.5 |
TANK
|
TRAF family member associated NFKB activator |
| chr12_+_57610150 | 0.03 |
ENST00000333972.11
|
ARHGEF25
|
Rho guanine nucleotide exchange factor 25 |
| chr2_+_233691607 | 0.03 |
ENST00000373424.5
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chr18_-_55321986 | 0.03 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr3_+_190388120 | 0.03 |
ENST00000456423.2
ENST00000264734.3 |
CLDN16
|
claudin 16 |
| chr1_+_15341744 | 0.03 |
ENST00000444385.5
|
FHAD1
|
forkhead associated phosphopeptide binding domain 1 |
| chr7_-_7535863 | 0.03 |
ENST00000399429.8
|
COL28A1
|
collagen type XXVIII alpha 1 chain |
| chr16_+_24537693 | 0.03 |
ENST00000564314.5
ENST00000567686.5 |
RBBP6
|
RB binding protein 6, ubiquitin ligase |
| chr12_+_53461015 | 0.02 |
ENST00000553064.6
ENST00000547859.2 |
PCBP2
|
poly(rC) binding protein 2 |
| chr3_-_98523013 | 0.02 |
ENST00000394181.6
ENST00000508902.5 ENST00000394180.6 |
CLDND1
|
claudin domain containing 1 |
| chr7_+_138460238 | 0.02 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr11_+_66011994 | 0.02 |
ENST00000312134.3
|
CST6
|
cystatin E/M |
| chr15_+_45252228 | 0.02 |
ENST00000560438.5
ENST00000347644.8 |
SLC28A2
|
solute carrier family 28 member 2 |
| chr5_+_36606355 | 0.02 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr1_+_108712903 | 0.02 |
ENST00000370017.9
|
FNDC7
|
fibronectin type III domain containing 7 |
| chr12_-_52553139 | 0.02 |
ENST00000267119.6
|
KRT71
|
keratin 71 |
| chr8_-_61689768 | 0.02 |
ENST00000517847.6
ENST00000389204.8 ENST00000517661.5 ENST00000517903.5 ENST00000522603.5 ENST00000541428.5 ENST00000522349.5 ENST00000522835.5 ENST00000518306.5 |
ASPH
|
aspartate beta-hydroxylase |
| chr12_-_10172117 | 0.02 |
ENST00000545927.5
ENST00000309539.8 ENST00000432556.6 ENST00000544577.5 |
OLR1
|
oxidized low density lipoprotein receptor 1 |
| chr6_-_26199272 | 0.02 |
ENST00000650491.1
ENST00000635200.1 ENST00000341023.2 |
ENSG00000282988.2
H2AC7
|
novel protein H2A clustered histone 7 |
| chrX_-_49002256 | 0.02 |
ENST00000611705.4
ENST00000376423.8 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chrX_-_49002197 | 0.02 |
ENST00000622231.1
ENST00000617369.4 ENST00000593475.5 ENST00000622599.4 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr9_+_74615582 | 0.02 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B |
| chr4_+_70195719 | 0.02 |
ENST00000683306.1
|
ODAM
|
odontogenic, ameloblast associated |
| chr7_+_90709530 | 0.02 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14 |
| chr18_-_55322215 | 0.02 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr2_-_31217511 | 0.02 |
ENST00000403897.4
|
CAPN14
|
calpain 14 |
| chr3_-_191282383 | 0.02 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr14_-_21098848 | 0.02 |
ENST00000556174.5
ENST00000554478.5 ENST00000553980.1 ENST00000421093.6 |
ZNF219
|
zinc finger protein 219 |
| chr12_+_71686473 | 0.02 |
ENST00000549735.5
|
TMEM19
|
transmembrane protein 19 |
| chr19_+_7030578 | 0.02 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3 like 5 |
| chr8_-_56211257 | 0.02 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chr6_+_112236806 | 0.02 |
ENST00000588837.5
ENST00000590293.5 ENST00000585450.5 ENST00000629766.2 ENST00000590804.5 ENST00000590584.4 ENST00000628122.2 ENST00000627025.1 ENST00000590673.5 ENST00000585611.5 ENST00000587816.2 |
LAMA4-AS1
ENSG00000281613.2
|
LAMA4 antisense RNA 1 novel ret finger protein-like 4B |
| chr2_-_70302048 | 0.02 |
ENST00000430566.6
ENST00000037869.7 |
FAM136A
|
family with sequence similarity 136 member A |
| chr5_+_146447304 | 0.02 |
ENST00000296702.9
ENST00000394421.7 ENST00000679501.2 |
TCERG1
|
transcription elongation regulator 1 |
| chr1_+_44213487 | 0.02 |
ENST00000315913.9
|
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr9_-_27005659 | 0.02 |
ENST00000380055.6
|
LRRC19
|
leucine rich repeat containing 19 |
| chr11_-_93197932 | 0.01 |
ENST00000326402.9
|
SLC36A4
|
solute carrier family 36 member 4 |
| chr7_+_90709231 | 0.01 |
ENST00000446790.5
ENST00000265741.7 |
CDK14
|
cyclin dependent kinase 14 |
| chr3_+_69936629 | 0.01 |
ENST00000394348.2
ENST00000531774.1 |
MITF
|
melanocyte inducing transcription factor |
| chr15_+_44427591 | 0.01 |
ENST00000558791.5
ENST00000260327.9 |
CTDSPL2
|
CTD small phosphatase like 2 |
| chr7_+_90709816 | 0.01 |
ENST00000436577.3
|
CDK14
|
cyclin dependent kinase 14 |
| chr1_+_44213440 | 0.01 |
ENST00000361745.10
ENST00000446292.5 ENST00000440641.5 ENST00000372289.7 ENST00000436069.5 ENST00000437511.5 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr3_+_69936583 | 0.01 |
ENST00000314557.10
ENST00000394351.9 |
MITF
|
melanocyte inducing transcription factor |
| chr3_-_196515315 | 0.01 |
ENST00000397537.3
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr6_-_32941018 | 0.01 |
ENST00000418107.3
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr12_-_7503744 | 0.01 |
ENST00000396620.7
ENST00000432237.3 |
CD163
|
CD163 molecule |
| chr18_+_10471829 | 0.01 |
ENST00000584596.2
|
APCDD1
|
APC down-regulated 1 |
| chr4_-_82844418 | 0.01 |
ENST00000503937.5
|
SEC31A
|
SEC31 homolog A, COPII coat complex component |
| chr17_-_41467386 | 0.01 |
ENST00000225899.4
|
KRT32
|
keratin 32 |
| chr3_+_12287859 | 0.01 |
ENST00000309576.11
ENST00000397015.7 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr3_+_12287899 | 0.01 |
ENST00000643888.2
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr14_-_80959484 | 0.01 |
ENST00000555529.5
ENST00000556042.5 ENST00000556981.5 |
CEP128
|
centrosomal protein 128 |
| chr3_+_154121366 | 0.01 |
ENST00000465093.6
ENST00000496710.5 ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor 26 |
| chr17_-_41124178 | 0.01 |
ENST00000394014.2
|
KRTAP4-12
|
keratin associated protein 4-12 |
| chr11_+_85647958 | 0.01 |
ENST00000304511.7
ENST00000528105.5 |
TMEM126A
|
transmembrane protein 126A |
| chr4_+_168092530 | 0.01 |
ENST00000359299.8
|
ANXA10
|
annexin A10 |
| chr4_-_75724362 | 0.00 |
ENST00000677583.1
|
G3BP2
|
G3BP stress granule assembly factor 2 |
| chr7_-_115968302 | 0.00 |
ENST00000457268.5
|
TFEC
|
transcription factor EC |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.1 | 0.5 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.4 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 0.4 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:0071284 | copper ion export(GO:0060003) cellular response to lead ion(GO:0071284) |
| 0.0 | 0.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:1904437 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0099404 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0072229 | proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.2 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.4 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:0014034 | neural crest cell fate commitment(GO:0014034) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.0 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.5 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |