Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for ALX3
Z-value: 0.49
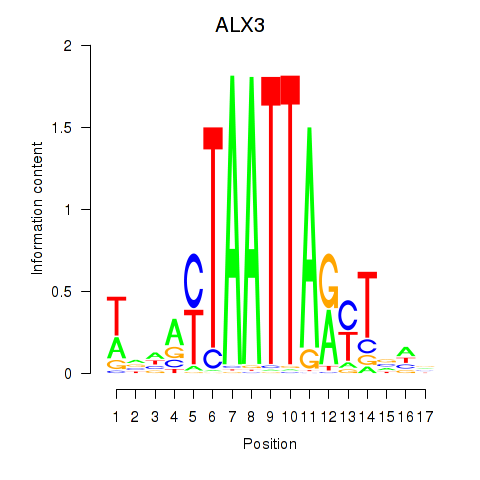
Transcription factors associated with ALX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ALX3
|
ENSG00000156150.9 | ALX3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ALX3 | hg38_v1_chr1_-_110070654_110070677 | 0.10 | 6.4e-01 | Click! |
Activity profile of ALX3 motif
Sorted Z-values of ALX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ALX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_85604146 | 1.29 |
ENST00000512201.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr6_+_127577168 | 1.03 |
ENST00000329722.8
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr2_-_224947030 | 0.76 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr6_+_26402237 | 0.59 |
ENST00000476549.6
ENST00000450085.6 ENST00000425234.6 ENST00000427334.5 ENST00000506698.1 ENST00000289361.11 |
BTN3A1
|
butyrophilin subfamily 3 member A1 |
| chr10_+_89392546 | 0.58 |
ENST00000546318.2
ENST00000371804.4 |
IFIT1
|
interferon induced protein with tetratricopeptide repeats 1 |
| chr12_+_26195647 | 0.46 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr2_+_102418642 | 0.45 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr6_+_26402289 | 0.44 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin subfamily 3 member A1 |
| chr20_+_33217325 | 0.42 |
ENST00000375452.3
ENST00000375454.8 |
BPIFA3
|
BPI fold containing family A member 3 |
| chr6_+_26440472 | 0.42 |
ENST00000494393.5
ENST00000482451.5 ENST00000471353.5 ENST00000361232.7 ENST00000487627.5 ENST00000496719.1 ENST00000244519.7 ENST00000490254.5 ENST00000487272.1 |
BTN3A3
|
butyrophilin subfamily 3 member A3 |
| chr8_-_85341705 | 0.41 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr12_+_26195313 | 0.39 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr1_-_150765785 | 0.39 |
ENST00000680311.1
ENST00000681728.1 ENST00000680288.1 |
CTSS
|
cathepsin S |
| chr8_+_49911801 | 0.38 |
ENST00000643809.1
|
SNTG1
|
syntrophin gamma 1 |
| chr17_+_1771688 | 0.37 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin family F member 1 |
| chr8_-_30812867 | 0.37 |
ENST00000518243.5
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta |
| chr5_-_35938572 | 0.37 |
ENST00000651391.1
ENST00000397366.5 ENST00000513623.5 ENST00000514524.2 ENST00000397367.6 |
CAPSL
|
calcyphosine like |
| chr15_+_44427793 | 0.35 |
ENST00000558966.5
ENST00000559793.5 ENST00000558968.1 |
CTDSPL2
|
CTD small phosphatase like 2 |
| chr8_-_115492221 | 0.34 |
ENST00000518018.1
|
TRPS1
|
transcriptional repressor GATA binding 1 |
| chr10_+_24466487 | 0.34 |
ENST00000396446.5
ENST00000396445.5 ENST00000376451.4 |
KIAA1217
|
KIAA1217 |
| chr2_-_219399981 | 0.34 |
ENST00000519905.1
ENST00000523282.5 ENST00000434339.5 ENST00000457935.5 |
DNPEP
|
aspartyl aminopeptidase |
| chr15_+_94355956 | 0.33 |
ENST00000557742.1
|
MCTP2
|
multiple C2 and transmembrane domain containing 2 |
| chr4_+_94974984 | 0.32 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr3_+_40477107 | 0.30 |
ENST00000314686.9
ENST00000447116.6 ENST00000429348.6 ENST00000432264.4 ENST00000456778.5 |
ZNF619
|
zinc finger protein 619 |
| chr14_+_34993240 | 0.30 |
ENST00000677647.1
|
SRP54
|
signal recognition particle 54 |
| chr2_+_68734861 | 0.30 |
ENST00000467265.5
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr12_+_26195543 | 0.29 |
ENST00000242729.7
|
SSPN
|
sarcospan |
| chr4_-_102828048 | 0.27 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr5_-_20575850 | 0.27 |
ENST00000507958.5
|
CDH18
|
cadherin 18 |
| chr15_+_64387828 | 0.27 |
ENST00000261884.8
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr18_+_31591869 | 0.26 |
ENST00000237014.8
|
TTR
|
transthyretin |
| chr2_+_102337148 | 0.26 |
ENST00000311734.6
ENST00000409584.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr11_+_35180279 | 0.26 |
ENST00000531873.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_-_230869564 | 0.26 |
ENST00000470540.5
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr4_+_94207596 | 0.26 |
ENST00000359052.8
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr7_-_111392915 | 0.25 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr8_+_106726115 | 0.25 |
ENST00000521592.5
|
OXR1
|
oxidation resistance 1 |
| chr11_+_92969651 | 0.25 |
ENST00000257068.3
ENST00000528076.1 |
MTNR1B
|
melatonin receptor 1B |
| chr12_-_23584600 | 0.24 |
ENST00000396007.6
|
SOX5
|
SRY-box transcription factor 5 |
| chr9_-_112332962 | 0.24 |
ENST00000458258.5
ENST00000210227.4 |
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr6_-_87095059 | 0.23 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr19_-_3557563 | 0.23 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr14_-_50561119 | 0.23 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr5_+_141382702 | 0.22 |
ENST00000617050.1
ENST00000518325.2 |
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr20_-_57690624 | 0.22 |
ENST00000414037.5
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr16_+_31259922 | 0.22 |
ENST00000648685.1
ENST00000544665.9 |
ITGAM
|
integrin subunit alpha M |
| chr14_-_106593319 | 0.22 |
ENST00000390627.3
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr1_-_23980345 | 0.21 |
ENST00000484146.6
|
SRSF10
|
serine and arginine rich splicing factor 10 |
| chr3_-_191282383 | 0.21 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr14_+_21997531 | 0.21 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr1_+_117001744 | 0.21 |
ENST00000256652.8
ENST00000682167.1 ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr10_-_114144599 | 0.20 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186 |
| chr17_-_74776323 | 0.20 |
ENST00000582870.5
ENST00000581136.5 ENST00000579218.5 ENST00000583476.5 ENST00000580301.5 ENST00000583757.5 ENST00000357814.8 ENST00000582524.5 |
NAT9
|
N-acetyltransferase 9 (putative) |
| chr12_+_1970772 | 0.20 |
ENST00000682544.1
|
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr10_+_35605337 | 0.19 |
ENST00000321660.2
|
GJD4
|
gap junction protein delta 4 |
| chr11_+_55827219 | 0.19 |
ENST00000378397.1
|
OR5L2
|
olfactory receptor family 5 subfamily L member 2 |
| chr12_+_1970809 | 0.19 |
ENST00000683781.1
ENST00000682462.1 |
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr11_-_62754141 | 0.19 |
ENST00000527994.1
ENST00000394807.5 ENST00000673933.1 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr6_+_160121859 | 0.19 |
ENST00000324965.8
ENST00000457470.6 |
SLC22A1
|
solute carrier family 22 member 1 |
| chr1_+_27935022 | 0.18 |
ENST00000411604.5
ENST00000373888.8 |
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr2_-_89085787 | 0.18 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr10_-_88851809 | 0.18 |
ENST00000371930.5
|
ANKRD22
|
ankyrin repeat domain 22 |
| chrX_+_83861126 | 0.18 |
ENST00000621735.4
ENST00000329312.5 |
CYLC1
|
cylicin 1 |
| chr4_-_102827723 | 0.17 |
ENST00000349311.12
|
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr8_-_30812773 | 0.17 |
ENST00000221138.9
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta |
| chr6_+_37929959 | 0.17 |
ENST00000373389.5
|
ZFAND3
|
zinc finger AN1-type containing 3 |
| chr3_-_112829367 | 0.17 |
ENST00000448932.4
ENST00000617549.3 |
CD200R1L
|
CD200 receptor 1 like |
| chr20_+_59019397 | 0.17 |
ENST00000217133.2
|
TUBB1
|
tubulin beta 1 class VI |
| chr17_-_66229380 | 0.17 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr3_+_119579577 | 0.16 |
ENST00000478927.5
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr1_+_27934980 | 0.16 |
ENST00000373894.8
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr4_-_102828022 | 0.16 |
ENST00000502690.5
|
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr2_+_68734773 | 0.16 |
ENST00000409202.8
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr14_+_32329256 | 0.16 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr2_+_90038848 | 0.16 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr1_-_247760556 | 0.16 |
ENST00000641256.1
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1 |
| chr11_+_6876625 | 0.15 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor family 10 subfamily A member 4 |
| chr1_-_92486916 | 0.15 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr11_-_61161414 | 0.15 |
ENST00000301765.10
|
VPS37C
|
VPS37C subunit of ESCRT-I |
| chr2_+_90172802 | 0.15 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr1_-_23980278 | 0.15 |
ENST00000374453.7
ENST00000453840.7 |
SRSF10
|
serine and arginine rich splicing factor 10 |
| chr20_+_34546815 | 0.15 |
ENST00000374837.7
|
MAP1LC3A
|
microtubule associated protein 1 light chain 3 alpha |
| chr11_-_122116215 | 0.15 |
ENST00000560104.2
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr3_+_119579676 | 0.15 |
ENST00000357003.7
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr1_+_27935110 | 0.14 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase acid like 3B |
| chr2_+_90234809 | 0.14 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr11_-_129024157 | 0.13 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr6_+_160121809 | 0.13 |
ENST00000366963.9
|
SLC22A1
|
solute carrier family 22 member 1 |
| chr10_+_27532521 | 0.13 |
ENST00000683924.1
|
RAB18
|
RAB18, member RAS oncogene family |
| chr3_+_122055355 | 0.13 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr6_-_169253835 | 0.13 |
ENST00000649844.1
ENST00000617924.6 |
THBS2
|
thrombospondin 2 |
| chr20_-_51802509 | 0.13 |
ENST00000371539.7
ENST00000217086.9 |
SALL4
|
spalt like transcription factor 4 |
| chr20_-_31390580 | 0.13 |
ENST00000339144.3
ENST00000376321.4 |
DEFB119
|
defensin beta 119 |
| chr9_-_96778053 | 0.13 |
ENST00000375231.5
ENST00000223428.9 |
ZNF510
|
zinc finger protein 510 |
| chr8_+_49911604 | 0.13 |
ENST00000642164.1
ENST00000644093.1 ENST00000643999.1 ENST00000647073.1 ENST00000646880.1 |
SNTG1
|
syntrophin gamma 1 |
| chr21_+_32298849 | 0.13 |
ENST00000303645.10
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chrX_+_108045050 | 0.13 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr11_-_16356538 | 0.12 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr15_+_44427591 | 0.12 |
ENST00000558791.5
ENST00000260327.9 |
CTDSPL2
|
CTD small phosphatase like 2 |
| chr10_+_47300174 | 0.12 |
ENST00000580279.2
|
GDF10
|
growth differentiation factor 10 |
| chr12_-_86256267 | 0.12 |
ENST00000620241.4
|
MGAT4C
|
MGAT4 family member C |
| chr6_-_7313146 | 0.12 |
ENST00000489567.5
ENST00000479365.5 ENST00000462112.1 ENST00000397511.6 ENST00000244763.9 ENST00000474597.5 ENST00000650389.1 |
SSR1
|
signal sequence receptor subunit 1 |
| chr4_+_109815503 | 0.12 |
ENST00000394631.7
|
GAR1
|
GAR1 ribonucleoprotein |
| chr14_+_22202561 | 0.12 |
ENST00000390460.1
|
TRAV26-2
|
T cell receptor alpha variable 26-2 |
| chr4_-_145180496 | 0.12 |
ENST00000447906.8
|
OTUD4
|
OTU deubiquitinase 4 |
| chr2_+_165239388 | 0.12 |
ENST00000424833.5
ENST00000375437.7 ENST00000631182.3 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr2_+_87338511 | 0.11 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr9_+_92947516 | 0.11 |
ENST00000375482.8
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr7_+_156640879 | 0.11 |
ENST00000311822.12
|
RNF32
|
ring finger protein 32 |
| chr8_-_673547 | 0.11 |
ENST00000522893.1
|
ERICH1
|
glutamate rich 1 |
| chr18_+_58341038 | 0.11 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chrX_+_1591590 | 0.11 |
ENST00000313871.9
ENST00000381261.8 |
AKAP17A
|
A-kinase anchoring protein 17A |
| chr17_-_41184895 | 0.11 |
ENST00000620667.1
ENST00000398472.2 |
KRTAP4-1
|
keratin associated protein 4-1 |
| chr4_+_87799546 | 0.11 |
ENST00000226284.7
|
IBSP
|
integrin binding sialoprotein |
| chr16_+_11965193 | 0.11 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr1_-_150697128 | 0.11 |
ENST00000427665.1
ENST00000271732.8 |
GOLPH3L
|
golgi phosphoprotein 3 like |
| chr17_+_18183052 | 0.10 |
ENST00000541285.1
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chrX_+_109535775 | 0.10 |
ENST00000218004.5
|
NXT2
|
nuclear transport factor 2 like export factor 2 |
| chr3_-_146528750 | 0.10 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1 |
| chr7_-_44541318 | 0.10 |
ENST00000381160.8
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr5_+_151259793 | 0.10 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr8_+_49911396 | 0.10 |
ENST00000642720.2
|
SNTG1
|
syntrophin gamma 1 |
| chr18_-_69956924 | 0.10 |
ENST00000581982.5
ENST00000280200.8 |
CD226
|
CD226 molecule |
| chrY_-_6872608 | 0.10 |
ENST00000383036.1
|
AMELY
|
amelogenin Y-linked |
| chr17_+_29941605 | 0.10 |
ENST00000394835.7
|
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr2_+_165239432 | 0.09 |
ENST00000636071.2
ENST00000636985.2 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr12_+_12357418 | 0.09 |
ENST00000298571.6
|
BORCS5
|
BLOC-1 related complex subunit 5 |
| chrX_+_108044967 | 0.09 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr2_-_101308681 | 0.09 |
ENST00000295317.4
|
RNF149
|
ring finger protein 149 |
| chr15_+_78873723 | 0.09 |
ENST00000559690.5
ENST00000559158.5 |
MORF4L1
|
mortality factor 4 like 1 |
| chr7_+_120988683 | 0.09 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr1_-_207052980 | 0.09 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr12_+_18261511 | 0.09 |
ENST00000538779.6
ENST00000433979.6 |
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr14_+_67720842 | 0.09 |
ENST00000267502.3
|
RDH12
|
retinol dehydrogenase 12 |
| chr11_-_107858777 | 0.08 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr2_-_206159194 | 0.08 |
ENST00000455934.6
ENST00000449699.5 ENST00000454195.1 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr19_-_14835162 | 0.08 |
ENST00000322301.5
|
OR7A5
|
olfactory receptor family 7 subfamily A member 5 |
| chr2_-_88979016 | 0.08 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr10_+_18260715 | 0.08 |
ENST00000615785.4
ENST00000617363.4 ENST00000396576.6 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr5_-_95081482 | 0.08 |
ENST00000312216.12
ENST00000512425.5 ENST00000505208.5 ENST00000429576.6 ENST00000508509.5 ENST00000510732.5 |
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr9_-_4666347 | 0.08 |
ENST00000381890.9
ENST00000682582.1 |
SPATA6L
|
spermatogenesis associated 6 like |
| chr11_+_72189528 | 0.08 |
ENST00000312293.9
|
FOLR1
|
folate receptor alpha |
| chr1_-_113871665 | 0.08 |
ENST00000528414.5
ENST00000460620.5 ENST00000359785.10 ENST00000420377.6 ENST00000525799.1 ENST00000538253.5 |
PTPN22
|
protein tyrosine phosphatase non-receptor type 22 |
| chr9_+_102995308 | 0.08 |
ENST00000612124.4
ENST00000374798.8 ENST00000487798.5 |
CYLC2
|
cylicin 2 |
| chr4_-_102828159 | 0.08 |
ENST00000394803.9
|
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr1_+_207325629 | 0.08 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr7_+_118224654 | 0.08 |
ENST00000265224.9
ENST00000486422.1 ENST00000417525.5 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr11_+_72189659 | 0.07 |
ENST00000393681.6
|
FOLR1
|
folate receptor alpha |
| chr16_-_28623560 | 0.07 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr9_+_69145463 | 0.07 |
ENST00000636438.1
|
TJP2
|
tight junction protein 2 |
| chr3_+_42979281 | 0.07 |
ENST00000488863.5
ENST00000430121.3 |
GASK1A
|
golgi associated kinase 1A |
| chr1_+_115029823 | 0.07 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta |
| chr4_-_69653223 | 0.07 |
ENST00000286604.8
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
|
UDP glucuronosyltransferase family 2 member A1 complex locus |
| chr15_+_58431985 | 0.07 |
ENST00000433326.2
ENST00000299022.10 |
LIPC
|
lipase C, hepatic type |
| chr6_+_154039333 | 0.07 |
ENST00000428397.6
|
OPRM1
|
opioid receptor mu 1 |
| chrX_-_152709260 | 0.07 |
ENST00000535353.3
ENST00000638741.1 ENST00000640702.1 |
CSAG2
|
CSAG family member 2 |
| chr5_+_140821598 | 0.07 |
ENST00000614258.1
ENST00000529859.2 ENST00000529619.5 |
PCDHA5
|
protocadherin alpha 5 |
| chr3_-_101320558 | 0.07 |
ENST00000193391.8
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr5_-_140346596 | 0.07 |
ENST00000230990.7
|
HBEGF
|
heparin binding EGF like growth factor |
| chr8_+_30387064 | 0.07 |
ENST00000523115.5
ENST00000519647.5 |
RBPMS
|
RNA binding protein, mRNA processing factor |
| chr11_+_55883297 | 0.06 |
ENST00000449290.6
|
TRIM51
|
tripartite motif-containing 51 |
| chr14_+_55027200 | 0.06 |
ENST00000395472.2
ENST00000555846.2 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr2_-_165203870 | 0.06 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr6_+_26183750 | 0.06 |
ENST00000614097.3
|
H2BC6
|
H2B clustered histone 6 |
| chr14_+_55027576 | 0.06 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr4_-_102827948 | 0.06 |
ENST00000394804.6
ENST00000394801.8 |
UBE2D3
|
ubiquitin conjugating enzyme E2 D3 |
| chr5_+_141177790 | 0.06 |
ENST00000239444.4
ENST00000623995.1 |
PCDHB8
ENSG00000279472.1
|
protocadherin beta 8 novel transcript |
| chr19_+_42076129 | 0.06 |
ENST00000359044.5
|
ZNF574
|
zinc finger protein 574 |
| chr4_-_119322128 | 0.06 |
ENST00000274024.4
|
FABP2
|
fatty acid binding protein 2 |
| chr9_+_12775012 | 0.06 |
ENST00000319264.4
|
LURAP1L
|
leucine rich adaptor protein 1 like |
| chr12_-_10130082 | 0.05 |
ENST00000533022.5
|
CLEC7A
|
C-type lectin domain containing 7A |
| chr7_-_87713287 | 0.05 |
ENST00000416177.1
ENST00000265724.8 ENST00000543898.5 |
ABCB1
|
ATP binding cassette subfamily B member 1 |
| chr16_-_28597042 | 0.05 |
ENST00000533150.5
ENST00000335715.9 |
SULT1A2
|
sulfotransferase family 1A member 2 |
| chr10_+_125896549 | 0.05 |
ENST00000368693.6
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr10_+_18400562 | 0.05 |
ENST00000377315.5
ENST00000650685.1 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr10_+_69269984 | 0.05 |
ENST00000436817.6
ENST00000450646.6 ENST00000360289.6 ENST00000448642.6 ENST00000464803.6 |
HK1
|
hexokinase 1 |
| chr1_-_23799561 | 0.05 |
ENST00000445705.1
|
GALE
|
UDP-galactose-4-epimerase |
| chr13_-_85799400 | 0.05 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr12_+_26011713 | 0.05 |
ENST00000542004.5
|
RASSF8
|
Ras association domain family member 8 |
| chr17_+_58169401 | 0.05 |
ENST00000641866.1
|
OR4D2
|
olfactory receptor family 4 subfamily D member 2 |
| chr20_-_51802433 | 0.05 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr17_+_18183803 | 0.05 |
ENST00000399138.5
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr16_-_55833085 | 0.05 |
ENST00000360526.8
|
CES1
|
carboxylesterase 1 |
| chr5_+_177384430 | 0.05 |
ENST00000512593.5
ENST00000324417.6 |
SLC34A1
|
solute carrier family 34 member 1 |
| chr16_-_28623330 | 0.04 |
ENST00000677940.1
|
ENSG00000288656.1
|
novel protein |
| chr14_+_39267377 | 0.04 |
ENST00000556148.5
ENST00000348007.7 |
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr11_-_62556230 | 0.04 |
ENST00000530285.5
|
AHNAK
|
AHNAK nucleoprotein |
| chr2_-_74392025 | 0.04 |
ENST00000440727.1
ENST00000409240.5 |
DCTN1
|
dynactin subunit 1 |
| chr18_+_45615473 | 0.04 |
ENST00000255226.11
|
SLC14A2
|
solute carrier family 14 member 2 |
| chr20_+_1118590 | 0.04 |
ENST00000246015.8
ENST00000335877.11 |
PSMF1
|
proteasome inhibitor subunit 1 |
| chr7_+_138460238 | 0.04 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr10_-_73655984 | 0.04 |
ENST00000394810.3
|
SYNPO2L
|
synaptopodin 2 like |
| chr7_-_44541262 | 0.04 |
ENST00000289547.8
ENST00000546276.5 ENST00000423141.1 |
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr8_+_104223344 | 0.04 |
ENST00000523362.5
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_+_224183197 | 0.04 |
ENST00000323699.9
|
DEGS1
|
delta 4-desaturase, sphingolipid 1 |
| chr6_-_82247697 | 0.04 |
ENST00000306270.12
ENST00000610980.4 |
IBTK
|
inhibitor of Bruton tyrosine kinase |
| chr1_+_157993273 | 0.04 |
ENST00000360089.8
ENST00000368173.7 |
KIRREL1
|
kirre like nephrin family adhesion molecule 1 |
| chr8_+_106726012 | 0.04 |
ENST00000449762.6
ENST00000297447.10 |
OXR1
|
oxidation resistance 1 |
| chr11_+_55811367 | 0.04 |
ENST00000625203.2
|
OR5L1
|
olfactory receptor family 5 subfamily L member 1 |
| chr9_+_108862255 | 0.04 |
ENST00000333999.5
|
ACTL7A
|
actin like 7A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.4 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.4 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.2 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.0 | 0.3 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0043016 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.0 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.5 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.4 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 1.1 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.3 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.0 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.1 | GO:0060368 | phosphatidylserine biosynthetic process(GO:0006659) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 1.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.3 | GO:0008513 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.3 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0030290 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |