|
chr12_+_112906777
Show fit
|
2.60 |
ENST00000452357.7
ENST00000445409.7
|
OAS1
|
2'-5'-oligoadenylate synthetase 1
|
|
chr1_+_78620722
Show fit
|
1.70 |
ENST00000679848.1
|
IFI44L
|
interferon induced protein 44 like
|
|
chr1_-_145996567
Show fit
|
1.63 |
ENST00000582401.6
|
TXNIP
|
thioredoxin interacting protein
|
|
chr4_+_70050431
Show fit
|
1.15 |
ENST00000511674.5
ENST00000246896.8
|
HTN1
|
histatin 1
|
|
chr12_-_70788914
Show fit
|
0.97 |
ENST00000342084.8
|
PTPRR
|
protein tyrosine phosphatase receptor type R
|
|
chr13_-_33205997
Show fit
|
0.94 |
ENST00000399365.7
|
STARD13
|
StAR related lipid transfer domain containing 13
|
|
chr10_+_89332484
Show fit
|
0.92 |
ENST00000371811.4
ENST00000680037.1
ENST00000679583.1
ENST00000679897.1
|
IFIT3
|
interferon induced protein with tetratricopeptide repeats 3
|
|
chr11_+_35180342
Show fit
|
0.83 |
ENST00000639002.1
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr12_+_96912517
Show fit
|
0.77 |
ENST00000457368.2
|
NEDD1
|
NEDD1 gamma-tubulin ring complex targeting factor
|
|
chr13_-_101416441
Show fit
|
0.77 |
ENST00000675332.1
ENST00000676315.1
ENST00000251127.11
|
NALCN
|
sodium leak channel, non-selective
|
|
chr11_+_35180279
Show fit
|
0.74 |
ENST00000531873.5
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr13_-_101416362
Show fit
|
0.71 |
ENST00000675802.1
ENST00000675150.1
|
NALCN
|
sodium leak channel, non-selective
|
|
chr4_-_80073170
Show fit
|
0.71 |
ENST00000403729.7
|
ANTXR2
|
ANTXR cell adhesion molecule 2
|
|
chr10_+_110005804
Show fit
|
0.71 |
ENST00000360162.7
|
ADD3
|
adducin 3
|
|
chr5_-_60488055
Show fit
|
0.70 |
ENST00000505507.6
ENST00000515835.2
ENST00000502484.6
|
PDE4D
|
phosphodiesterase 4D
|
|
chr10_-_122845850
Show fit
|
0.67 |
ENST00000392790.6
|
CUZD1
|
CUB and zona pellucida like domains 1
|
|
chr8_-_63026179
Show fit
|
0.64 |
ENST00000677919.1
|
GGH
|
gamma-glutamyl hydrolase
|
|
chr1_-_89126066
Show fit
|
0.62 |
ENST00000370466.4
|
GBP2
|
guanylate binding protein 2
|
|
chr4_-_80073057
Show fit
|
0.60 |
ENST00000681710.1
|
ANTXR2
|
ANTXR cell adhesion molecule 2
|
|
chr20_-_18413216
Show fit
|
0.58 |
ENST00000480488.2
|
DZANK1
|
double zinc ribbon and ankyrin repeat domains 1
|
|
chr11_-_105035113
Show fit
|
0.58 |
ENST00000526568.5
ENST00000531166.5
ENST00000534497.5
ENST00000527979.5
ENST00000533400.6
ENST00000528974.1
ENST00000525825.5
ENST00000353247.9
ENST00000446369.5
ENST00000436863.7
|
CASP1
|
caspase 1
|
|
chr1_-_154627906
Show fit
|
0.58 |
ENST00000679899.1
|
ADAR
|
adenosine deaminase RNA specific
|
|
chr2_+_118942188
Show fit
|
0.58 |
ENST00000327097.5
|
MARCO
|
macrophage receptor with collagenous structure
|
|
chr1_-_154627945
Show fit
|
0.56 |
ENST00000681683.1
ENST00000368471.8
ENST00000649042.1
ENST00000680270.1
ENST00000649022.2
ENST00000681056.1
ENST00000649724.1
|
ADAR
|
adenosine deaminase RNA specific
|
|
chr15_+_64387828
Show fit
|
0.56 |
ENST00000261884.8
|
TRIP4
|
thyroid hormone receptor interactor 4
|
|
chr18_+_616711
Show fit
|
0.56 |
ENST00000579494.1
|
CLUL1
|
clusterin like 1
|
|
chr4_-_80073465
Show fit
|
0.54 |
ENST00000404191.5
|
ANTXR2
|
ANTXR cell adhesion molecule 2
|
|
chr12_+_64497968
Show fit
|
0.53 |
ENST00000676593.1
ENST00000677093.1
|
TBK1
ENSG00000288665.1
|
TANK binding kinase 1
novel transcript
|
|
chr18_+_616672
Show fit
|
0.53 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1
|
|
chr10_-_99235783
Show fit
|
0.52 |
ENST00000370546.5
ENST00000614306.1
|
HPSE2
|
heparanase 2 (inactive)
|
|
chr8_+_12104389
Show fit
|
0.51 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D
|
|
chr15_-_58014097
Show fit
|
0.50 |
ENST00000559517.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2
|
|
chr3_-_52770856
Show fit
|
0.50 |
ENST00000461689.5
ENST00000535191.5
ENST00000383721.8
ENST00000233027.10
|
NEK4
|
NIMA related kinase 4
|
|
chr10_-_99235846
Show fit
|
0.49 |
ENST00000370552.8
ENST00000370549.5
ENST00000628193.2
|
HPSE2
|
heparanase 2 (inactive)
|
|
chr17_-_69244846
Show fit
|
0.48 |
ENST00000269081.8
|
ABCA10
|
ATP binding cassette subfamily A member 10
|
|
chr3_+_142723999
Show fit
|
0.46 |
ENST00000476941.6
ENST00000273482.10
|
TRPC1
|
transient receptor potential cation channel subfamily C member 1
|
|
chr1_-_54764482
Show fit
|
0.45 |
ENST00000371279.4
|
PARS2
|
prolyl-tRNA synthetase 2, mitochondrial
|
|
chr9_+_116153783
Show fit
|
0.45 |
ENST00000328252.4
|
PAPPA
|
pappalysin 1
|
|
chr4_-_68670648
Show fit
|
0.43 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15
|
|
chr11_+_71538025
Show fit
|
0.43 |
ENST00000398534.4
|
KRTAP5-8
|
keratin associated protein 5-8
|
|
chr11_+_45804038
Show fit
|
0.43 |
ENST00000442528.2
ENST00000526817.1
|
SLC35C1
|
solute carrier family 35 member C1
|
|
chr5_+_162067858
Show fit
|
0.43 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2
|
|
chr16_-_72094371
Show fit
|
0.41 |
ENST00000426362.6
|
TXNL4B
|
thioredoxin like 4B
|
|
chr4_-_122621011
Show fit
|
0.39 |
ENST00000611104.2
ENST00000648588.1
|
IL21
|
interleukin 21
|
|
chr8_-_61646807
Show fit
|
0.39 |
ENST00000522919.5
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr9_-_19786928
Show fit
|
0.38 |
ENST00000341998.6
ENST00000286344.3
|
SLC24A2
|
solute carrier family 24 member 2
|
|
chr6_-_29087313
Show fit
|
0.38 |
ENST00000377173.4
|
OR2B3
|
olfactory receptor family 2 subfamily B member 3
|
|
chrX_+_141894666
Show fit
|
0.38 |
ENST00000544766.5
|
MAGEC3
|
MAGE family member C3
|
|
chr5_+_162067990
Show fit
|
0.38 |
ENST00000641017.1
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2
|
|
chr2_+_167187364
Show fit
|
0.38 |
ENST00000672671.1
|
XIRP2
|
xin actin binding repeat containing 2
|
|
chr4_+_73436198
Show fit
|
0.37 |
ENST00000395792.7
|
AFP
|
alpha fetoprotein
|
|
chr2_+_105851748
Show fit
|
0.37 |
ENST00000425756.1
ENST00000393349.2
|
NCK2
|
NCK adaptor protein 2
|
|
chr3_-_191282383
Show fit
|
0.37 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B
|
|
chr20_+_59604527
Show fit
|
0.36 |
ENST00000371015.6
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr4_+_112647059
Show fit
|
0.36 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein 7, transcriptional regulator
|
|
chr18_-_54357820
Show fit
|
0.36 |
ENST00000577499.5
ENST00000584040.1
ENST00000581310.5
|
STARD6
|
StAR related lipid transfer domain containing 6
|
|
chr4_+_70028452
Show fit
|
0.35 |
ENST00000530128.5
ENST00000381057.3
ENST00000673563.1
|
HTN3
|
histatin 3
|
|
chr12_-_39340963
Show fit
|
0.35 |
ENST00000552961.5
|
KIF21A
|
kinesin family member 21A
|
|
chr4_+_73436244
Show fit
|
0.35 |
ENST00000226359.2
|
AFP
|
alpha fetoprotein
|
|
chr6_+_4889992
Show fit
|
0.34 |
ENST00000343762.5
|
CDYL
|
chromodomain Y like
|
|
chr2_-_135047432
Show fit
|
0.34 |
ENST00000392915.7
ENST00000637841.1
ENST00000414343.1
|
MAP3K19
|
mitogen-activated protein kinase kinase kinase 19
|
|
chr2_-_89117844
Show fit
|
0.34 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17
|
|
chr3_+_46354072
Show fit
|
0.33 |
ENST00000445132.3
ENST00000421659.1
|
CCR2
|
C-C motif chemokine receptor 2
|
|
chr4_-_168318770
Show fit
|
0.33 |
ENST00000680771.1
ENST00000514995.2
|
DDX60
|
DExD/H-box helicase 60
|
|
chr10_+_116545907
Show fit
|
0.32 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase
|
|
chr4_-_168318743
Show fit
|
0.32 |
ENST00000393743.8
|
DDX60
|
DExD/H-box helicase 60
|
|
chr6_+_132570322
Show fit
|
0.32 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6
|
|
chr17_+_18781250
Show fit
|
0.31 |
ENST00000476139.5
|
TVP23B
|
trans-golgi network vesicle protein 23 homolog B
|
|
chr2_-_216694794
Show fit
|
0.31 |
ENST00000449583.1
|
IGFBP5
|
insulin like growth factor binding protein 5
|
|
chr3_+_173398438
Show fit
|
0.31 |
ENST00000457714.5
|
NLGN1
|
neuroligin 1
|
|
chr2_+_234050732
Show fit
|
0.31 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2
|
|
chr19_+_20776292
Show fit
|
0.30 |
ENST00000360204.5
ENST00000344519.10
ENST00000594534.5
|
ZNF66
|
zinc finger protein 66
|
|
chr19_-_53254841
Show fit
|
0.30 |
ENST00000601828.5
ENST00000599012.5
ENST00000598513.6
ENST00000598806.5
|
ZNF677
|
zinc finger protein 677
|
|
chr2_-_162152404
Show fit
|
0.30 |
ENST00000375497.3
|
GCG
|
glucagon
|
|
chr10_+_89283685
Show fit
|
0.30 |
ENST00000638108.1
|
IFIT2
|
interferon induced protein with tetratricopeptide repeats 2
|
|
chr12_+_75480800
Show fit
|
0.30 |
ENST00000456650.7
|
GLIPR1
|
GLI pathogenesis related 1
|
|
chr8_+_103880412
Show fit
|
0.30 |
ENST00000436393.6
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr17_-_31314066
Show fit
|
0.30 |
ENST00000577894.1
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr5_+_90899183
Show fit
|
0.30 |
ENST00000640815.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1
|
|
chr8_+_18391276
Show fit
|
0.29 |
ENST00000286479.4
ENST00000520116.1
|
NAT2
|
N-acetyltransferase 2
|
|
chr12_+_133181409
Show fit
|
0.29 |
ENST00000416488.5
ENST00000228289.9
ENST00000541211.6
ENST00000536435.7
ENST00000500625.7
ENST00000539248.6
ENST00000542711.6
ENST00000536899.6
ENST00000542986.6
ENST00000611984.4
ENST00000541975.2
|
ZNF268
|
zinc finger protein 268
|
|
chr17_+_18781155
Show fit
|
0.28 |
ENST00000574226.5
ENST00000575261.5
ENST00000307767.13
|
TVP23B
|
trans-golgi network vesicle protein 23 homolog B
|
|
chr1_-_205422050
Show fit
|
0.28 |
ENST00000367153.9
|
LEMD1
|
LEM domain containing 1
|
|
chr3_-_132684685
Show fit
|
0.28 |
ENST00000512094.5
ENST00000632629.1
|
NPHP3
NPHP3-ACAD11
|
nephrocystin 3
NPHP3-ACAD11 readthrough (NMD candidate)
|
|
chr3_-_121660892
Show fit
|
0.28 |
ENST00000428394.6
ENST00000314583.8
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr11_+_73950985
Show fit
|
0.28 |
ENST00000339764.6
|
DNAJB13
|
DnaJ heat shock protein family (Hsp40) member B13
|
|
chr11_-_86672419
Show fit
|
0.28 |
ENST00000524826.7
ENST00000532471.1
|
ME3
|
malic enzyme 3
|
|
chr2_+_109794296
Show fit
|
0.28 |
ENST00000430736.5
ENST00000016946.8
ENST00000441344.1
|
RGPD5
|
RANBP2 like and GRIP domain containing 5
|
|
chr12_+_75481204
Show fit
|
0.27 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis related 1
|
|
chr3_+_169911566
Show fit
|
0.27 |
ENST00000428432.6
ENST00000335556.7
|
SAMD7
|
sterile alpha motif domain containing 7
|
|
chr19_-_48110793
Show fit
|
0.27 |
ENST00000599111.5
ENST00000599921.6
|
PLA2G4C
|
phospholipase A2 group IVC
|
|
chr12_+_75480745
Show fit
|
0.27 |
ENST00000266659.8
|
GLIPR1
|
GLI pathogenesis related 1
|
|
chr1_-_247104278
Show fit
|
0.27 |
ENST00000366501.1
ENST00000366500.5
ENST00000448299.7
ENST00000476158.2
ENST00000343381.10
|
ZNF669
|
zinc finger protein 669
|
|
chr10_+_68109433
Show fit
|
0.27 |
ENST00000613327.4
ENST00000358913.10
ENST00000373675.3
|
MYPN
|
myopalladin
|
|
chr19_-_48110775
Show fit
|
0.27 |
ENST00000354276.7
|
PLA2G4C
|
phospholipase A2 group IVC
|
|
chr12_-_53207271
Show fit
|
0.27 |
ENST00000552972.5
ENST00000422257.7
|
ITGB7
|
integrin subunit beta 7
|
|
chr9_-_114657791
Show fit
|
0.26 |
ENST00000423632.3
|
TEX53
|
testis expressed 53
|
|
chr10_-_27154226
Show fit
|
0.26 |
ENST00000427324.5
ENST00000375972.7
ENST00000326799.7
|
YME1L1
|
YME1 like 1 ATPase
|
|
chr3_+_98463201
Show fit
|
0.26 |
ENST00000642057.1
|
OR5K1
|
olfactory receptor family 5 subfamily K member 1
|
|
chr4_-_67883987
Show fit
|
0.25 |
ENST00000283916.11
|
TMPRSS11D
|
transmembrane serine protease 11D
|
|
chr7_-_73842508
Show fit
|
0.25 |
ENST00000297873.9
|
METTL27
|
methyltransferase like 27
|
|
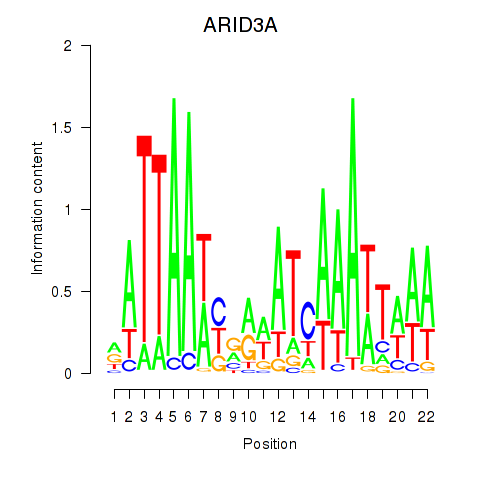
chr19_+_926001
Show fit
|
0.25 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A
|
|
chr7_+_141776674
Show fit
|
0.25 |
ENST00000247881.4
|
TAS2R4
|
taste 2 receptor member 4
|
|
chr1_-_20119527
Show fit
|
0.25 |
ENST00000375105.8
ENST00000617227.1
|
PLA2G2D
|
phospholipase A2 group IID
|
|
chr17_+_3207539
Show fit
|
0.25 |
ENST00000641322.1
ENST00000641732.2
|
OR1A1
|
olfactory receptor family 1 subfamily A member 1
|
|
chr4_+_188139438
Show fit
|
0.25 |
ENST00000332517.4
|
TRIML1
|
tripartite motif family like 1
|
|
chr16_-_48247533
Show fit
|
0.25 |
ENST00000356608.7
ENST00000569991.1
|
ABCC11
|
ATP binding cassette subfamily C member 11
|
|
chr4_+_174918355
Show fit
|
0.24 |
ENST00000505141.5
ENST00000359240.7
ENST00000615367.4
ENST00000445694.5
ENST00000618444.1
|
ADAM29
|
ADAM metallopeptidase domain 29
|
|
chr1_+_156126160
Show fit
|
0.24 |
ENST00000448611.6
ENST00000368297.5
|
LMNA
|
lamin A/C
|
|
chr6_-_46735693
Show fit
|
0.24 |
ENST00000537365.1
|
PLA2G7
|
phospholipase A2 group VII
|
|
chr14_-_106269133
Show fit
|
0.24 |
ENST00000390609.3
|
IGHV3-23
|
immunoglobulin heavy variable 3-23
|
|
chr2_+_90154073
Show fit
|
0.23 |
ENST00000611391.1
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13
|
|
chr12_-_53207241
Show fit
|
0.23 |
ENST00000267082.10
ENST00000549086.2
|
ITGB7
|
integrin subunit beta 7
|
|
chr19_+_35748549
Show fit
|
0.23 |
ENST00000301159.14
|
LIN37
|
lin-37 DREAM MuvB core complex component
|
|
chr17_-_15563428
Show fit
|
0.23 |
ENST00000584811.5
ENST00000419890.3
ENST00000518321.6
ENST00000438826.7
ENST00000225576.7
ENST00000428082.6
ENST00000522212.6
|
TVP23C
TVP23C-CDRT4
|
trans-golgi network vesicle protein 23 homolog C
TVP23C-CDRT4 readthrough
|
|
chr8_+_32721823
Show fit
|
0.23 |
ENST00000539990.3
ENST00000519240.5
|
NRG1
|
neuregulin 1
|
|
chr9_-_21351378
Show fit
|
0.23 |
ENST00000380210.1
|
IFNA6
|
interferon alpha 6
|
|
chr7_-_22500152
Show fit
|
0.23 |
ENST00000406890.6
ENST00000678116.1
ENST00000424363.5
|
STEAP1B
|
STEAP family member 1B
|
|
chr6_-_30213379
Show fit
|
0.23 |
ENST00000418026.1
ENST00000454678.7
ENST00000416596.5
ENST00000453195.5
|
TRIM26
|
tripartite motif containing 26
|
|
chr12_+_10505602
Show fit
|
0.22 |
ENST00000322446.3
|
EIF2S3B
|
eukaryotic translation initiation factor 2 subunit gamma B
|
|
chr19_-_12830655
Show fit
|
0.22 |
ENST00000586969.5
ENST00000589681.5
ENST00000585384.5
ENST00000393233.6
ENST00000589808.5
|
RTBDN
|
retbindin
|
|
chr3_-_94028582
Show fit
|
0.22 |
ENST00000315099.3
|
STX19
|
syntaxin 19
|
|
chr7_+_76461676
Show fit
|
0.22 |
ENST00000425780.5
ENST00000456590.5
ENST00000451769.5
ENST00000324432.9
ENST00000457529.5
ENST00000446600.5
ENST00000430490.7
ENST00000413936.6
ENST00000423646.5
ENST00000438930.5
|
DTX2
|
deltex E3 ubiquitin ligase 2
|
|
chr18_+_32091849
Show fit
|
0.22 |
ENST00000261593.8
ENST00000578914.1
|
RNF138
|
ring finger protein 138
|
|
chr11_-_86672630
Show fit
|
0.22 |
ENST00000543262.5
|
ME3
|
malic enzyme 3
|
|
chr11_-_84720876
Show fit
|
0.21 |
ENST00000648622.1
|
DLG2
|
discs large MAGUK scaffold protein 2
|
|
chr11_-_1608463
Show fit
|
0.21 |
ENST00000399685.1
|
KRTAP5-3
|
keratin associated protein 5-3
|
|
chr16_-_4767125
Show fit
|
0.20 |
ENST00000219478.11
ENST00000545009.1
|
ZNF500
|
zinc finger protein 500
|
|
chr10_-_67838173
Show fit
|
0.20 |
ENST00000225171.7
|
DNAJC12
|
DnaJ heat shock protein family (Hsp40) member C12
|
|
chr18_-_13915531
Show fit
|
0.20 |
ENST00000327606.4
|
MC2R
|
melanocortin 2 receptor
|
|
chr16_+_70646536
Show fit
|
0.20 |
ENST00000288098.6
|
IL34
|
interleukin 34
|
|
chr7_-_122699108
Show fit
|
0.20 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133
|
|
chr3_-_71493500
Show fit
|
0.19 |
ENST00000648380.1
ENST00000650295.1
|
FOXP1
|
forkhead box P1
|
|
chr16_+_56936654
Show fit
|
0.19 |
ENST00000563911.5
|
HERPUD1
|
homocysteine inducible ER protein with ubiquitin like domain 1
|
|
chr4_+_15339818
Show fit
|
0.19 |
ENST00000397700.6
ENST00000295297.4
|
C1QTNF7
|
C1q and TNF related 7
|
|
chr3_+_119173564
Show fit
|
0.19 |
ENST00000264234.8
ENST00000479520.5
ENST00000494855.5
|
UPK1B
|
uroplakin 1B
|
|
chr13_-_46897021
Show fit
|
0.19 |
ENST00000542664.4
ENST00000543956.5
|
HTR2A
|
5-hydroxytryptamine receptor 2A
|
|
chr1_+_211326615
Show fit
|
0.19 |
ENST00000336184.6
|
TRAF5
|
TNF receptor associated factor 5
|
|
chr20_-_43726989
Show fit
|
0.18 |
ENST00000373003.2
|
GTSF1L
|
gametocyte specific factor 1 like
|
|
chr9_-_111484353
Show fit
|
0.18 |
ENST00000338205.9
ENST00000684092.1
|
ECPAS
|
Ecm29 proteasome adaptor and scaffold
|
|
chr12_+_10505890
Show fit
|
0.18 |
ENST00000538173.1
|
EIF2S3B
|
eukaryotic translation initiation factor 2 subunit gamma B
|
|
chr16_-_21857657
Show fit
|
0.18 |
ENST00000341400.11
ENST00000518761.8
ENST00000682606.1
|
NPIPB4
|
nuclear pore complex interacting protein family member B4
|
|
chr17_-_73232218
Show fit
|
0.18 |
ENST00000583024.1
ENST00000403627.7
ENST00000405159.7
ENST00000581110.1
|
FAM104A
|
family with sequence similarity 104 member A
|
|
chr9_-_21482313
Show fit
|
0.17 |
ENST00000448696.4
|
IFNE
|
interferon epsilon
|
|
chr1_+_171557845
Show fit
|
0.17 |
ENST00000644916.1
|
PRRC2C
|
proline rich coiled-coil 2C
|
|
chr5_-_137499293
Show fit
|
0.17 |
ENST00000510689.5
ENST00000394945.6
|
SPOCK1
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 1
|
|
chr12_+_25973748
Show fit
|
0.17 |
ENST00000542865.5
|
RASSF8
|
Ras association domain family member 8
|
|
chr12_+_32679200
Show fit
|
0.17 |
ENST00000452533.6
ENST00000414834.6
|
DNM1L
|
dynamin 1 like
|
|
chr14_+_39265278
Show fit
|
0.17 |
ENST00000554392.5
ENST00000555716.5
ENST00000341749.7
ENST00000557038.5
|
MIA2
|
MIA SH3 domain ER export factor 2
|
|
chr16_+_19410480
Show fit
|
0.17 |
ENST00000541464.5
|
TMC5
|
transmembrane channel like 5
|
|
chr16_+_7510102
Show fit
|
0.17 |
ENST00000682918.1
ENST00000683326.1
ENST00000570626.2
|
RBFOX1
|
RNA binding fox-1 homolog 1
|
|
chr18_+_63887698
Show fit
|
0.17 |
ENST00000457692.5
ENST00000299502.9
ENST00000413956.5
|
SERPINB2
|
serpin family B member 2
|
|
chr10_+_26438317
Show fit
|
0.17 |
ENST00000376236.9
|
APBB1IP
|
amyloid beta precursor protein binding family B member 1 interacting protein
|
|
chr19_+_3762665
Show fit
|
0.17 |
ENST00000330133.5
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr14_-_75051426
Show fit
|
0.17 |
ENST00000556257.5
ENST00000355774.7
ENST00000557648.1
ENST00000553263.1
ENST00000380968.6
|
MLH3
|
mutL homolog 3
|
|
chr16_-_21857418
Show fit
|
0.17 |
ENST00000415645.6
|
NPIPB4
|
nuclear pore complex interacting protein family member B4
|
|
chrX_+_1268786
Show fit
|
0.17 |
ENST00000501036.7
ENST00000417535.7
|
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha
|
|
chr12_+_119334722
Show fit
|
0.17 |
ENST00000327554.3
|
CCDC60
|
coiled-coil domain containing 60
|
|
chr22_+_20965108
Show fit
|
0.16 |
ENST00000399167.6
ENST00000399163.6
|
AIFM3
|
apoptosis inducing factor mitochondria associated 3
|
|
chr19_+_48695952
Show fit
|
0.16 |
ENST00000522966.2
ENST00000425340.3
ENST00000391876.5
|
FUT2
|
fucosyltransferase 2
|
|
chr1_-_72100930
Show fit
|
0.16 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1
|
|
chr10_+_5364955
Show fit
|
0.16 |
ENST00000380433.5
|
UCN3
|
urocortin 3
|
|
chr17_+_73232601
Show fit
|
0.16 |
ENST00000359042.6
|
C17orf80
|
chromosome 17 open reading frame 80
|
|
chr12_+_54498766
Show fit
|
0.16 |
ENST00000545638.2
|
NCKAP1L
|
NCK associated protein 1 like
|
|
chr17_+_7627963
Show fit
|
0.16 |
ENST00000575729.5
ENST00000340624.9
|
SHBG
|
sex hormone binding globulin
|
|
chr19_-_13506408
Show fit
|
0.15 |
ENST00000637736.1
ENST00000637432.1
ENST00000638029.1
ENST00000360228.11
ENST00000638009.2
ENST00000637769.1
ENST00000635895.1
|
CACNA1A
|
calcium voltage-gated channel subunit alpha1 A
|
|
chr2_-_108989206
Show fit
|
0.15 |
ENST00000258443.7
ENST00000409271.5
ENST00000376651.1
|
EDAR
|
ectodysplasin A receptor
|
|
chr16_-_11281322
Show fit
|
0.15 |
ENST00000312511.4
|
PRM1
|
protamine 1
|
|
chr14_-_68794597
Show fit
|
0.15 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein like 1
|
|
chr8_+_84183534
Show fit
|
0.15 |
ENST00000518566.5
|
RALYL
|
RALY RNA binding protein like
|
|
chrX_-_15315615
Show fit
|
0.15 |
ENST00000380470.7
ENST00000480796.6
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chrX_+_1268807
Show fit
|
0.15 |
ENST00000381524.8
ENST00000381529.9
ENST00000412290.6
|
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha
|
|
chr12_+_32679269
Show fit
|
0.15 |
ENST00000358214.9
ENST00000553257.6
ENST00000549701.6
ENST00000266481.10
ENST00000551476.5
ENST00000550154.5
ENST00000547312.5
ENST00000381000.8
ENST00000548750.5
|
DNM1L
|
dynamin 1 like
|
|
chr6_+_52420992
Show fit
|
0.15 |
ENST00000636954.1
ENST00000636566.1
ENST00000638075.1
|
EFHC1
|
EF-hand domain containing 1
|
|
chr4_+_56978877
Show fit
|
0.15 |
ENST00000433463.1
ENST00000314595.6
|
POLR2B
|
RNA polymerase II subunit B
|
|
chr16_-_29404029
Show fit
|
0.15 |
ENST00000524087.5
|
NPIPB11
|
nuclear pore complex interacting protein family member B11
|
|
chr7_-_38265678
Show fit
|
0.15 |
ENST00000443402.6
|
TRGC1
|
T cell receptor gamma constant 1
|
|
chr12_-_10802627
Show fit
|
0.14 |
ENST00000240687.2
|
TAS2R7
|
taste 2 receptor member 7
|
|
chr1_-_94925759
Show fit
|
0.14 |
ENST00000415017.1
ENST00000545882.5
|
CNN3
|
calponin 3
|
|
chr4_+_56978858
Show fit
|
0.14 |
ENST00000431623.6
ENST00000441246.6
|
POLR2B
|
RNA polymerase II subunit B
|
|
chr13_+_97960192
Show fit
|
0.14 |
ENST00000496368.6
ENST00000421861.7
ENST00000357602.7
|
IPO5
|
importin 5
|
|
chr9_-_83956677
Show fit
|
0.14 |
ENST00000376344.8
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr7_-_124929938
Show fit
|
0.14 |
ENST00000668382.1
ENST00000655761.1
ENST00000393329.5
|
POT1
|
protection of telomeres 1
|
|
chr2_-_110577101
Show fit
|
0.14 |
ENST00000330331.9
ENST00000446930.1
ENST00000329516.8
|
RGPD6
|
RANBP2 like and GRIP domain containing 6
|
|
chr6_-_116060859
Show fit
|
0.14 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase
|
|
chr12_-_4379712
Show fit
|
0.14 |
ENST00000237837.2
|
FGF23
|
fibroblast growth factor 23
|
|
chr22_+_23856703
Show fit
|
0.14 |
ENST00000345044.10
|
SLC2A11
|
solute carrier family 2 member 11
|
|
chr4_+_70397931
Show fit
|
0.14 |
ENST00000399575.7
|
OPRPN
|
opiorphin prepropeptide
|
|
chr2_-_70190900
Show fit
|
0.14 |
ENST00000425268.5
ENST00000428751.5
ENST00000417203.5
ENST00000417865.5
ENST00000428010.5
ENST00000447804.1
ENST00000264434.7
|
C2orf42
|
chromosome 2 open reading frame 42
|
|
chr16_-_21278282
Show fit
|
0.14 |
ENST00000572914.2
|
CRYM
|
crystallin mu
|
|
chrX_-_20141810
Show fit
|
0.14 |
ENST00000379593.1
ENST00000379607.10
|
EIF1AX
|
eukaryotic translation initiation factor 1A X-linked
|
|
chr19_-_44401575
Show fit
|
0.14 |
ENST00000591679.5
ENST00000544719.6
ENST00000614994.5
ENST00000585868.1
ENST00000588212.1
|
ZNF285
ENSG00000267173.1
|
zinc finger protein 285
novel protein
|
|
chr16_+_22513523
Show fit
|
0.14 |
ENST00000538606.5
ENST00000451409.5
ENST00000424340.5
ENST00000517539.5
ENST00000528249.5
|
NPIPB5
|
nuclear pore complex interacting protein family member B5
|
|
chr21_+_32298849
Show fit
|
0.14 |
ENST00000303645.10
|
MRAP
|
melanocortin 2 receptor accessory protein
|
|
chr14_+_19743571
Show fit
|
0.14 |
ENST00000642117.2
|
OR4Q3
|
olfactory receptor family 4 subfamily Q member 3
|
|
chr14_+_23630109
Show fit
|
0.13 |
ENST00000432832.6
|
DHRS2
|
dehydrogenase/reductase 2
|
|
chr4_-_103077282
Show fit
|
0.13 |
ENST00000503230.5
ENST00000503818.1
|
SLC9B2
|
solute carrier family 9 member B2
|
|
chr12_+_69239560
Show fit
|
0.13 |
ENST00000435070.7
|
CPSF6
|
cleavage and polyadenylation specific factor 6
|
|
chr6_+_54083423
Show fit
|
0.13 |
ENST00000460844.6
ENST00000370876.6
|
MLIP
|
muscular LMNA interacting protein
|
|
chr16_+_30053123
Show fit
|
0.13 |
ENST00000395248.6
ENST00000575627.5
ENST00000566897.6
ENST00000568435.6
ENST00000338110.11
ENST00000562240.1
|
ENSG00000285043.3
|
novel protein
|