Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for ARID5A
Z-value: 0.72
Transcription factors associated with ARID5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5A
|
ENSG00000196843.17 | ARID5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5A | hg38_v1_chr2_+_96536743_96536762 | 0.69 | 1.2e-04 | Click! |
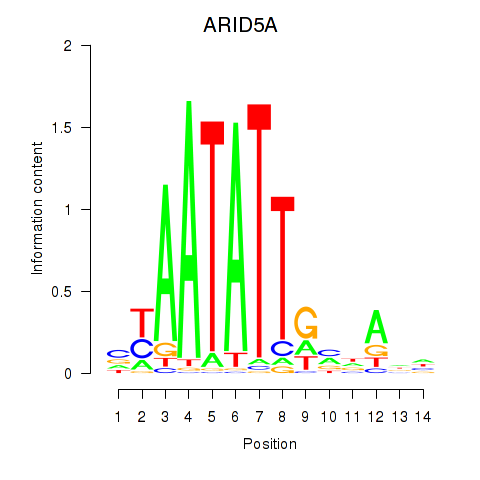
Activity profile of ARID5A motif
Sorted Z-values of ARID5A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_151357583 | 5.76 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6 |
| chr5_+_119354771 | 3.72 |
ENST00000503646.1
|
TNFAIP8
|
TNF alpha induced protein 8 |
| chr1_-_169734064 | 2.13 |
ENST00000333360.12
|
SELE
|
selectin E |
| chr9_-_120926752 | 1.94 |
ENST00000373887.8
|
TRAF1
|
TNF receptor associated factor 1 |
| chr2_-_224947030 | 1.64 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr1_-_89065200 | 1.58 |
ENST00000370473.5
|
GBP1
|
guanylate binding protein 1 |
| chr1_-_89126066 | 1.58 |
ENST00000370466.4
|
GBP2
|
guanylate binding protein 2 |
| chr6_+_29099490 | 1.51 |
ENST00000641659.2
|
OR2J1
|
olfactory receptor family 2 subfamily J member 1 |
| chr6_-_10415043 | 1.51 |
ENST00000379613.10
|
TFAP2A
|
transcription factor AP-2 alpha |
| chr14_+_22070548 | 1.43 |
ENST00000390450.3
|
TRAV22
|
T cell receptor alpha variable 22 |
| chr16_-_75556214 | 1.37 |
ENST00000568377.5
ENST00000565067.5 ENST00000258173.11 |
TMEM231
|
transmembrane protein 231 |
| chr12_+_77830886 | 1.35 |
ENST00000397909.7
ENST00000549464.5 |
NAV3
|
neuron navigator 3 |
| chr1_-_173050931 | 1.34 |
ENST00000404377.5
|
TNFSF18
|
TNF superfamily member 18 |
| chr2_-_174846405 | 1.33 |
ENST00000409597.5
ENST00000413882.6 |
CHN1
|
chimerin 1 |
| chr7_-_22193824 | 1.29 |
ENST00000401957.6
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr2_-_174847015 | 1.24 |
ENST00000650938.1
|
CHN1
|
chimerin 1 |
| chr18_+_63887698 | 1.23 |
ENST00000457692.5
ENST00000299502.9 ENST00000413956.5 |
SERPINB2
|
serpin family B member 2 |
| chr6_-_10414985 | 1.18 |
ENST00000466073.5
ENST00000498450.3 |
TFAP2A
|
transcription factor AP-2 alpha |
| chr8_+_85177225 | 1.12 |
ENST00000418930.6
|
E2F5
|
E2F transcription factor 5 |
| chr21_-_34526850 | 1.08 |
ENST00000481448.5
ENST00000381132.6 |
RCAN1
|
regulator of calcineurin 1 |
| chr2_+_102418642 | 1.01 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr2_+_102337148 | 0.98 |
ENST00000311734.6
ENST00000409584.5 |
IL1RL1
|
interleukin 1 receptor like 1 |
| chr4_-_68670648 | 0.92 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr2_-_24328113 | 0.91 |
ENST00000622089.4
|
ITSN2
|
intersectin 2 |
| chr18_+_58149314 | 0.75 |
ENST00000435432.6
ENST00000357895.9 ENST00000586263.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr15_+_70936487 | 0.73 |
ENST00000558456.5
ENST00000560158.6 ENST00000558808.5 ENST00000559806.5 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr3_+_11154477 | 0.71 |
ENST00000431010.3
|
HRH1
|
histamine receptor H1 |
| chr4_-_176195563 | 0.68 |
ENST00000280191.7
|
SPATA4
|
spermatogenesis associated 4 |
| chr1_-_154627945 | 0.64 |
ENST00000681683.1
ENST00000368471.8 ENST00000649042.1 ENST00000680270.1 ENST00000649022.2 ENST00000681056.1 ENST00000649724.1 |
ADAR
|
adenosine deaminase RNA specific |
| chr12_+_9827472 | 0.64 |
ENST00000617793.4
ENST00000617889.5 ENST00000354855.7 ENST00000279545.7 |
KLRF1
|
killer cell lectin like receptor F1 |
| chr6_-_169253835 | 0.64 |
ENST00000649844.1
ENST00000617924.6 |
THBS2
|
thrombospondin 2 |
| chr5_+_126423363 | 0.63 |
ENST00000285689.8
|
GRAMD2B
|
GRAM domain containing 2B |
| chr18_+_31591869 | 0.62 |
ENST00000237014.8
|
TTR
|
transthyretin |
| chr16_-_48247533 | 0.60 |
ENST00000356608.7
ENST00000569991.1 |
ABCC11
|
ATP binding cassette subfamily C member 11 |
| chr12_+_69825221 | 0.60 |
ENST00000552032.7
|
MYRFL
|
myelin regulatory factor like |
| chr19_-_43670153 | 0.59 |
ENST00000601723.5
ENST00000339082.7 ENST00000340093.8 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr5_+_55024250 | 0.55 |
ENST00000231009.3
|
GZMK
|
granzyme K |
| chr6_+_46793379 | 0.54 |
ENST00000230588.9
ENST00000611727.2 |
MEP1A
|
meprin A subunit alpha |
| chr17_-_3063607 | 0.54 |
ENST00000575751.1
|
OR1D5
|
olfactory receptor family 1 subfamily D member 5 |
| chr11_+_55883297 | 0.51 |
ENST00000449290.6
|
TRIM51
|
tripartite motif-containing 51 |
| chr1_+_40988513 | 0.51 |
ENST00000649215.1
|
CTPS1
|
CTP synthase 1 |
| chr10_+_110005804 | 0.50 |
ENST00000360162.7
|
ADD3
|
adducin 3 |
| chr1_-_154627906 | 0.50 |
ENST00000679899.1
|
ADAR
|
adenosine deaminase RNA specific |
| chr19_+_33374312 | 0.50 |
ENST00000585933.2
|
CEBPG
|
CCAAT enhancer binding protein gamma |
| chr12_+_112125531 | 0.49 |
ENST00000549358.5
ENST00000257604.9 ENST00000548092.5 ENST00000412615.7 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chrX_+_48897919 | 0.48 |
ENST00000447146.7
ENST00000247140.8 |
PQBP1
|
polyglutamine binding protein 1 |
| chr21_-_34526815 | 0.48 |
ENST00000492600.1
|
RCAN1
|
regulator of calcineurin 1 |
| chr6_+_10528326 | 0.47 |
ENST00000379597.7
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr11_+_60429595 | 0.46 |
ENST00000528905.1
ENST00000528093.1 |
MS4A5
|
membrane spanning 4-domains A5 |
| chr3_+_101827982 | 0.46 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr3_+_9649433 | 0.46 |
ENST00000353332.9
ENST00000420925.5 ENST00000296003.9 ENST00000351233.9 |
MTMR14
|
myotubularin related protein 14 |
| chr1_+_86468902 | 0.45 |
ENST00000394711.2
|
CLCA1
|
chloride channel accessory 1 |
| chr19_+_54502799 | 0.45 |
ENST00000301202.7
|
LAIR2
|
leukocyte associated immunoglobulin like receptor 2 |
| chr2_+_167135901 | 0.44 |
ENST00000628543.2
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr19_+_54502867 | 0.42 |
ENST00000351841.2
|
LAIR2
|
leukocyte associated immunoglobulin like receptor 2 |
| chr11_-_5324297 | 0.41 |
ENST00000624187.1
|
OR51B2
|
olfactory receptor family 51 subfamily B member 2 |
| chr1_-_89270751 | 0.41 |
ENST00000370459.7
|
GBP5
|
guanylate binding protein 5 |
| chr1_+_116909869 | 0.41 |
ENST00000393203.3
|
PTGFRN
|
prostaglandin F2 receptor inhibitor |
| chr7_+_16661182 | 0.41 |
ENST00000446596.5
ENST00000452975.6 ENST00000438834.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr3_+_97439603 | 0.41 |
ENST00000514100.5
|
EPHA6
|
EPH receptor A6 |
| chr10_+_24466487 | 0.40 |
ENST00000396446.5
ENST00000396445.5 ENST00000376451.4 |
KIAA1217
|
KIAA1217 |
| chr12_+_21131187 | 0.40 |
ENST00000256958.3
|
SLCO1B1
|
solute carrier organic anion transporter family member 1B1 |
| chr4_-_70666492 | 0.40 |
ENST00000254801.9
ENST00000391614.7 |
JCHAIN
|
joining chain of multimeric IgA and IgM |
| chr18_+_58362467 | 0.40 |
ENST00000675101.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_-_100178215 | 0.39 |
ENST00000370138.1
ENST00000370137.6 ENST00000342895.7 ENST00000620882.4 |
LRRC39
|
leucine rich repeat containing 39 |
| chr12_+_69825273 | 0.38 |
ENST00000547771.6
|
MYRFL
|
myelin regulatory factor like |
| chr1_-_212847649 | 0.38 |
ENST00000332912.3
|
SPATA45
|
spermatogenesis associated 45 |
| chr19_+_35748549 | 0.38 |
ENST00000301159.14
|
LIN37
|
lin-37 DREAM MuvB core complex component |
| chr11_-_48983826 | 0.37 |
ENST00000649162.1
|
TRIM51GP
|
tripartite motif-containing 51G, pseudogene |
| chr3_-_150703943 | 0.37 |
ENST00000491361.5
|
ERICH6
|
glutamate rich 6 |
| chr3_+_148730100 | 0.36 |
ENST00000474935.5
ENST00000475347.5 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor type 1 |
| chr2_+_102001966 | 0.36 |
ENST00000457817.5
|
IL1R2
|
interleukin 1 receptor type 2 |
| chr3_-_151278535 | 0.36 |
ENST00000309170.8
|
P2RY14
|
purinergic receptor P2Y14 |
| chr2_-_10447771 | 0.36 |
ENST00000405333.5
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr4_-_47981535 | 0.35 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr2_-_179861604 | 0.35 |
ENST00000410066.7
|
ZNF385B
|
zinc finger protein 385B |
| chr4_+_54229261 | 0.35 |
ENST00000508170.5
ENST00000512143.1 ENST00000257290.10 |
PDGFRA
|
platelet derived growth factor receptor alpha |
| chrX_+_80420466 | 0.34 |
ENST00000308293.5
|
TENT5D
|
terminal nucleotidyltransferase 5D |
| chr6_-_26271815 | 0.34 |
ENST00000614378.1
|
H3C8
|
H3 clustered histone 8 |
| chr6_-_49744378 | 0.33 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr6_-_49744434 | 0.33 |
ENST00000433368.6
ENST00000354620.4 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr6_+_30067530 | 0.33 |
ENST00000376765.6
ENST00000376763.5 |
PPP1R11
|
protein phosphatase 1 regulatory inhibitor subunit 11 |
| chr1_-_197067234 | 0.32 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr1_+_248231417 | 0.32 |
ENST00000641868.1
|
OR2M4
|
olfactory receptor family 2 subfamily M member 4 |
| chrX_+_48897900 | 0.32 |
ENST00000376566.8
ENST00000376563.5 |
PQBP1
|
polyglutamine binding protein 1 |
| chr19_+_3224701 | 0.32 |
ENST00000541430.6
|
CELF5
|
CUGBP Elav-like family member 5 |
| chr7_+_116953306 | 0.32 |
ENST00000265437.9
ENST00000393451.7 |
ST7
|
suppression of tumorigenicity 7 |
| chr3_+_75906666 | 0.32 |
ENST00000487694.7
ENST00000602589.5 |
ROBO2
|
roundabout guidance receptor 2 |
| chr21_-_18403754 | 0.32 |
ENST00000284885.8
|
TMPRSS15
|
transmembrane serine protease 15 |
| chr2_-_73779968 | 0.32 |
ENST00000452812.1
ENST00000443070.5 ENST00000272444.7 |
DUSP11
|
dual specificity phosphatase 11 |
| chr11_-_31810991 | 0.32 |
ENST00000640684.1
|
PAX6
|
paired box 6 |
| chr8_+_36784324 | 0.31 |
ENST00000523973.5
ENST00000399881.8 |
KCNU1
|
potassium calcium-activated channel subfamily U member 1 |
| chr9_-_92482350 | 0.31 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr4_+_186227501 | 0.31 |
ENST00000446598.6
ENST00000264690.11 ENST00000513864.2 |
KLKB1
|
kallikrein B1 |
| chr6_+_10555787 | 0.31 |
ENST00000316170.9
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr9_-_34381578 | 0.31 |
ENST00000379133.7
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr5_-_132543513 | 0.31 |
ENST00000231454.6
|
IL5
|
interleukin 5 |
| chr2_+_89936859 | 0.31 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr6_-_49866453 | 0.30 |
ENST00000507853.5
|
CRISP1
|
cysteine rich secretory protein 1 |
| chr12_+_10307950 | 0.30 |
ENST00000543420.5
ENST00000543777.5 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr20_+_31475278 | 0.30 |
ENST00000201979.3
|
REM1
|
RRAD and GEM like GTPase 1 |
| chr7_-_32071397 | 0.29 |
ENST00000396184.7
ENST00000396189.2 ENST00000321453.12 |
PDE1C
|
phosphodiesterase 1C |
| chr12_-_124989053 | 0.29 |
ENST00000308736.7
|
DHX37
|
DEAH-box helicase 37 |
| chrX_+_83861126 | 0.29 |
ENST00000621735.4
ENST00000329312.5 |
CYLC1
|
cylicin 1 |
| chr1_+_12857086 | 0.29 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr5_-_170389349 | 0.29 |
ENST00000274629.9
|
KCNMB1
|
potassium calcium-activated channel subfamily M regulatory beta subunit 1 |
| chr10_+_69088096 | 0.28 |
ENST00000242465.4
|
SRGN
|
serglycin |
| chrX_+_154762729 | 0.28 |
ENST00000620277.4
|
DKC1
|
dyskerin pseudouridine synthase 1 |
| chr11_-_6030758 | 0.28 |
ENST00000641900.1
|
OR56A1
|
olfactory receptor family 56 subfamily A member 1 |
| chr9_-_34381531 | 0.27 |
ENST00000379124.5
ENST00000379126.7 ENST00000379127.1 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chr6_-_152563271 | 0.27 |
ENST00000535896.7
ENST00000672122.1 |
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr1_+_21977014 | 0.27 |
ENST00000337107.11
|
CELA3B
|
chymotrypsin like elastase 3B |
| chr14_-_74875998 | 0.27 |
ENST00000556489.4
ENST00000673765.1 |
PROX2
|
prospero homeobox 2 |
| chrX_+_65588368 | 0.27 |
ENST00000609672.5
|
MSN
|
moesin |
| chr18_+_31376777 | 0.27 |
ENST00000308128.9
ENST00000359747.4 |
DSG4
|
desmoglein 4 |
| chr11_-_31811231 | 0.27 |
ENST00000438681.6
|
PAX6
|
paired box 6 |
| chr10_-_60572599 | 0.26 |
ENST00000503366.5
|
ANK3
|
ankyrin 3 |
| chr12_-_16600703 | 0.26 |
ENST00000616247.4
|
LMO3
|
LIM domain only 3 |
| chr8_-_86069662 | 0.26 |
ENST00000276616.3
|
PSKH2
|
protein serine kinase H2 |
| chr7_+_16646131 | 0.26 |
ENST00000415365.5
ENST00000433922.6 ENST00000630952.2 ENST00000258761.8 ENST00000405202.5 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr11_-_102724945 | 0.26 |
ENST00000236826.8
|
MMP8
|
matrix metallopeptidase 8 |
| chr3_+_14675128 | 0.26 |
ENST00000435614.5
ENST00000253697.8 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr8_-_28490220 | 0.25 |
ENST00000517673.5
ENST00000380254.7 ENST00000518734.5 ENST00000346498.6 |
FBXO16
|
F-box protein 16 |
| chr1_+_170935526 | 0.25 |
ENST00000367758.7
ENST00000367759.9 |
MROH9
|
maestro heat like repeat family member 9 |
| chr20_+_38962299 | 0.25 |
ENST00000373325.6
ENST00000373323.8 ENST00000252011.8 ENST00000615559.1 |
DHX35
ENSG00000277581.1
|
DEAH-box helicase 35 novel transcript, sense intronic to DHX35 |
| chr2_-_227379297 | 0.25 |
ENST00000304568.4
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr5_+_141370236 | 0.25 |
ENST00000576222.2
ENST00000618934.1 |
PCDHGB3
|
protocadherin gamma subfamily B, 3 |
| chr16_+_30748241 | 0.24 |
ENST00000565924.5
ENST00000424889.7 |
PHKG2
|
phosphorylase kinase catalytic subunit gamma 2 |
| chr11_+_61228377 | 0.24 |
ENST00000537932.5
|
PGA4
|
pepsinogen A4 |
| chr3_+_73061659 | 0.24 |
ENST00000533473.1
|
EBLN2
|
endogenous Bornavirus like nucleoprotein 2 |
| chr6_+_116399395 | 0.24 |
ENST00000644499.1
|
ENSG00000285446.1
|
novel protein |
| chr19_-_43820598 | 0.24 |
ENST00000594049.5
ENST00000414615.6 |
LYPD5
|
LY6/PLAUR domain containing 5 |
| chr18_-_46964408 | 0.24 |
ENST00000676383.1
|
ENSG00000288616.1
|
elongin A3 family member D |
| chr2_-_212124901 | 0.24 |
ENST00000402597.6
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4 |
| chr14_+_90256542 | 0.24 |
ENST00000261303.13
ENST00000543772.2 |
PSMC1
|
proteasome 26S subunit, ATPase 1 |
| chr19_+_10013468 | 0.24 |
ENST00000591589.3
|
RDH8
|
retinol dehydrogenase 8 |
| chr20_+_142573 | 0.23 |
ENST00000382398.4
|
DEFB126
|
defensin beta 126 |
| chr4_+_183905266 | 0.23 |
ENST00000308497.9
|
STOX2
|
storkhead box 2 |
| chr6_-_26056460 | 0.23 |
ENST00000343677.4
|
H1-2
|
H1.2 linker histone, cluster member |
| chr19_+_35641728 | 0.23 |
ENST00000619399.4
ENST00000379026.6 ENST00000379023.8 ENST00000402764.6 ENST00000479824.5 |
ETV2
|
ETS variant transcription factor 2 |
| chr15_+_94297939 | 0.23 |
ENST00000357742.9
|
MCTP2
|
multiple C2 and transmembrane domain containing 2 |
| chr15_-_72231583 | 0.23 |
ENST00000566809.1
ENST00000567087.5 ENST00000569050.1 ENST00000568883.5 |
PKM
|
pyruvate kinase M1/2 |
| chr6_-_49866527 | 0.23 |
ENST00000335847.9
|
CRISP1
|
cysteine rich secretory protein 1 |
| chrX_+_41724174 | 0.23 |
ENST00000302548.5
|
GPR82
|
G protein-coupled receptor 82 |
| chr19_+_54497879 | 0.23 |
ENST00000412608.5
ENST00000610651.1 |
LAIR2
|
leukocyte associated immunoglobulin like receptor 2 |
| chr4_+_70383123 | 0.22 |
ENST00000304915.8
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr9_+_113444725 | 0.22 |
ENST00000374140.6
|
RGS3
|
regulator of G protein signaling 3 |
| chr10_+_96000091 | 0.22 |
ENST00000424464.5
ENST00000410012.6 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr19_-_2292024 | 0.22 |
ENST00000585527.1
|
LINGO3
|
leucine rich repeat and Ig domain containing 3 |
| chr14_+_56117702 | 0.22 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr12_+_25958891 | 0.22 |
ENST00000381352.7
ENST00000535907.5 ENST00000405154.6 ENST00000615708.4 |
RASSF8
|
Ras association domain family member 8 |
| chrX_+_22032301 | 0.22 |
ENST00000379374.5
|
PHEX
|
phosphate regulating endopeptidase homolog X-linked |
| chr7_+_134843884 | 0.22 |
ENST00000445569.6
|
CALD1
|
caldesmon 1 |
| chr5_-_58999885 | 0.21 |
ENST00000317118.12
|
PDE4D
|
phosphodiesterase 4D |
| chr5_+_141387698 | 0.21 |
ENST00000615384.1
ENST00000519479.2 |
PCDHGB4
|
protocadherin gamma subfamily B, 4 |
| chr3_+_171843337 | 0.21 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr1_+_117420597 | 0.21 |
ENST00000449370.6
|
MAN1A2
|
mannosidase alpha class 1A member 2 |
| chr3_-_87276577 | 0.20 |
ENST00000344265.8
ENST00000350375.7 |
POU1F1
|
POU class 1 homeobox 1 |
| chr4_+_69931066 | 0.20 |
ENST00000246891.9
|
CSN1S1
|
casein alpha s1 |
| chr12_+_9971402 | 0.20 |
ENST00000304361.9
ENST00000396507.7 ENST00000434319.6 |
CLEC12A
|
C-type lectin domain family 12 member A |
| chr19_-_893172 | 0.20 |
ENST00000325464.6
ENST00000312090.10 |
MED16
|
mediator complex subunit 16 |
| chr9_+_107306459 | 0.20 |
ENST00000457811.1
|
RAD23B
|
RAD23 homolog B, nucleotide excision repair protein |
| chr4_+_118850668 | 0.19 |
ENST00000610556.4
|
SYNPO2
|
synaptopodin 2 |
| chr6_-_130222813 | 0.19 |
ENST00000437477.6
ENST00000439090.7 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr11_-_31811034 | 0.19 |
ENST00000638250.1
|
PAX6
|
paired box 6 |
| chr11_-_31811112 | 0.19 |
ENST00000640242.1
ENST00000640613.1 ENST00000606377.7 |
PAX6
|
paired box 6 |
| chr4_+_67558719 | 0.19 |
ENST00000265404.7
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr5_+_58491451 | 0.19 |
ENST00000513924.2
ENST00000515443.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr3_+_178558700 | 0.19 |
ENST00000432997.5
ENST00000455865.5 |
KCNMB2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr1_+_158845798 | 0.19 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr5_+_141397942 | 0.19 |
ENST00000617380.2
ENST00000621169.1 |
PCDHGB5
|
protocadherin gamma subfamily B, 5 |
| chr5_+_58491427 | 0.19 |
ENST00000396776.6
ENST00000502276.6 ENST00000511930.2 |
GAPT
|
GRB2 binding adaptor protein, transmembrane |
| chr4_+_109912877 | 0.19 |
ENST00000265171.10
ENST00000509793.5 ENST00000652245.1 |
EGF
|
epidermal growth factor |
| chr13_-_110242694 | 0.18 |
ENST00000648989.1
ENST00000647797.1 ENST00000648966.1 ENST00000649484.1 ENST00000648695.1 ENST00000650115.1 ENST00000650566.1 |
COL4A1
|
collagen type IV alpha 1 chain |
| chr11_-_31811140 | 0.18 |
ENST00000639916.1
|
PAX6
|
paired box 6 |
| chrX_+_86714623 | 0.18 |
ENST00000484479.1
|
DACH2
|
dachshund family transcription factor 2 |
| chr4_+_70226116 | 0.18 |
ENST00000317987.6
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr10_+_46375645 | 0.18 |
ENST00000622769.4
|
ANXA8L1
|
annexin A8 like 1 |
| chr3_-_167474026 | 0.18 |
ENST00000466903.1
ENST00000264677.8 |
SERPINI2
|
serpin family I member 2 |
| chr10_+_46375619 | 0.18 |
ENST00000584982.7
ENST00000613703.4 |
ANXA8L1
|
annexin A8 like 1 |
| chr8_-_51809414 | 0.18 |
ENST00000356297.5
|
PXDNL
|
peroxidasin like |
| chr10_+_27532521 | 0.18 |
ENST00000683924.1
|
RAB18
|
RAB18, member RAS oncogene family |
| chr1_-_66801276 | 0.18 |
ENST00000304526.3
|
INSL5
|
insulin like 5 |
| chr10_+_94683722 | 0.18 |
ENST00000285979.11
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr2_-_32264853 | 0.18 |
ENST00000402280.6
|
NLRC4
|
NLR family CARD domain containing 4 |
| chr22_+_25219633 | 0.18 |
ENST00000398215.3
|
CRYBB2
|
crystallin beta B2 |
| chr11_-_114595777 | 0.17 |
ENST00000375478.4
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr14_+_67720842 | 0.17 |
ENST00000267502.3
|
RDH12
|
retinol dehydrogenase 12 |
| chr6_+_122996227 | 0.17 |
ENST00000275162.10
|
CLVS2
|
clavesin 2 |
| chr6_+_30328889 | 0.17 |
ENST00000396547.5
ENST00000623385.3 |
TRIM39
TRIM39-RPP21
|
tripartite motif containing 39 TRIM39-RPP21 readthrough |
| chrX_-_101386166 | 0.17 |
ENST00000308731.8
ENST00000372880.5 |
BTK
|
Bruton tyrosine kinase |
| chr10_+_133394094 | 0.17 |
ENST00000477902.6
|
MTG1
|
mitochondrial ribosome associated GTPase 1 |
| chr11_+_89924064 | 0.17 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr19_+_7030578 | 0.16 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3 like 5 |
| chr13_+_42781578 | 0.16 |
ENST00000313851.3
|
FAM216B
|
family with sequence similarity 216 member B |
| chr1_+_171185293 | 0.16 |
ENST00000209929.10
|
FMO2
|
flavin containing dimethylaniline monoxygenase 2 |
| chr11_-_5269933 | 0.16 |
ENST00000396895.3
|
HBE1
|
hemoglobin subunit epsilon 1 |
| chr4_-_99352730 | 0.16 |
ENST00000510055.5
ENST00000515683.6 ENST00000511397.3 |
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide |
| chr2_-_68952880 | 0.16 |
ENST00000481498.1
ENST00000328895.9 |
GKN2
|
gastrokine 2 |
| chr13_+_42781547 | 0.16 |
ENST00000537894.5
|
FAM216B
|
family with sequence similarity 216 member B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.3 | 1.3 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.3 | 1.6 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.2 | 0.6 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.2 | 1.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.2 | 5.8 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.4 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 1.0 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.9 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 | 0.3 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.1 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.3 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 1.6 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 1.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.4 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.3 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 | 2.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.2 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 1.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:0051919 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 1.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.1 | 0.4 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.3 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.2 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.1 | 0.3 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.1 | 0.2 | GO:0002368 | positive regulation of type I hypersensitivity(GO:0001812) B cell selection(GO:0002339) B cell cytokine production(GO:0002368) |
| 0.1 | 0.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.4 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 1.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 2.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.4 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.1 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.5 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 1.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.3 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:1900127 | negative regulation of cholesterol efflux(GO:0090370) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:1904021 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.0 | 1.6 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.6 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 3.7 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.1 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.5 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.0 | 2.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.8 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.4 | GO:0071756 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.1 | 0.3 | GO:0033150 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.1 | 6.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.2 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.1 | 0.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.2 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.3 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 1.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 2.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 1.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 2.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 1.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 5.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 1.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.3 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.1 | 0.5 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 3.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.4 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.3 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.4 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.6 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 1.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 2.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 4.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 1.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 1.3 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 2.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.9 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 6.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 2.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |