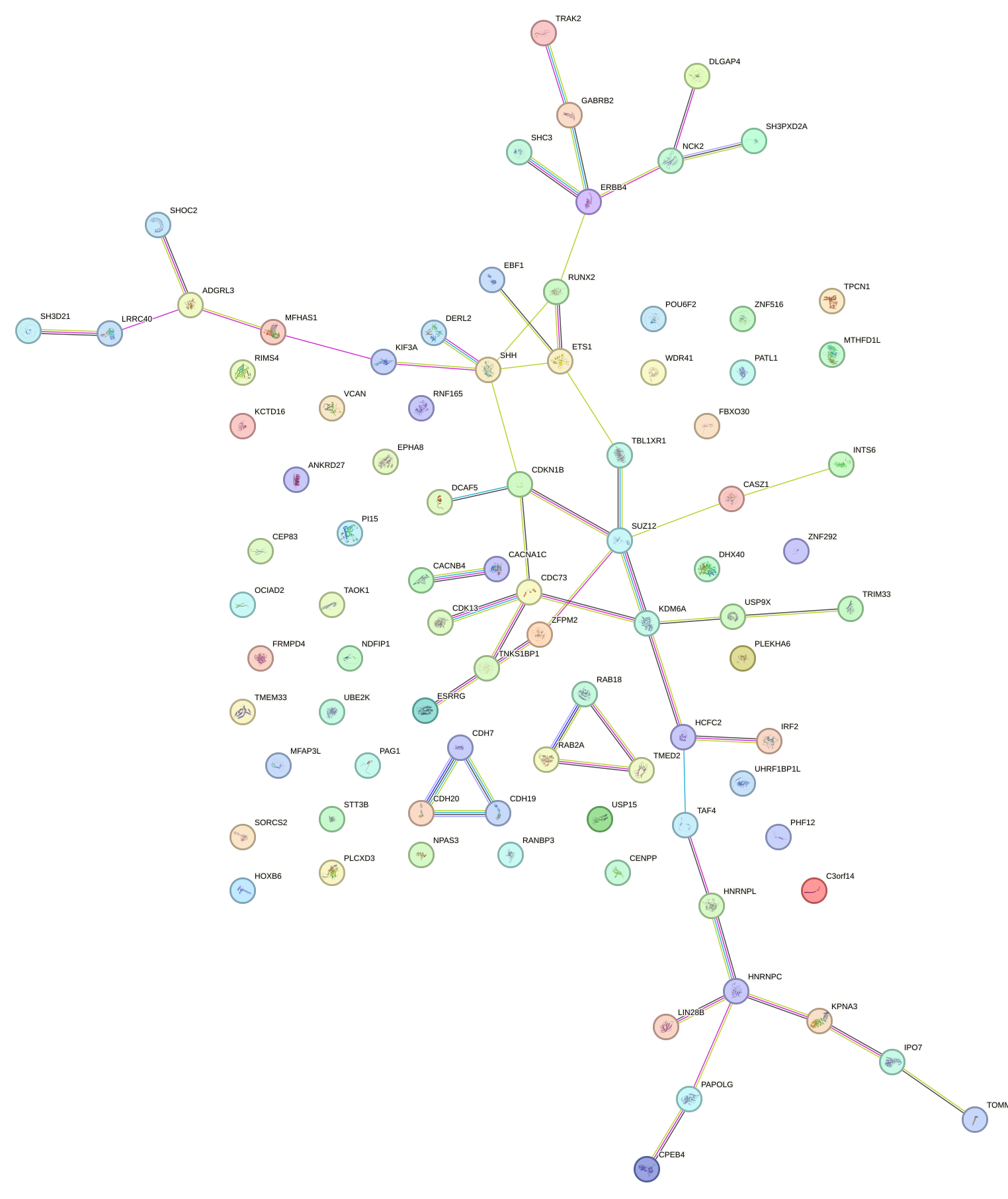
|
chr10_-_103855406
Show fit
|
0.34 |
ENST00000355946.6
ENST00000369774.8
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr8_+_105318428
Show fit
|
0.16 |
ENST00000407775.7
|
ZFPM2
|
zinc finger protein, FOG family member 2
|
|
chr4_-_184474518
Show fit
|
0.15 |
ENST00000393593.8
|
IRF2
|
interferon regulatory factor 2
|
|
chr11_-_128522264
Show fit
|
0.12 |
ENST00000531611.5
|
ETS1
|
ETS proto-oncogene 1, transcription factor
|
|
chr5_-_77492309
Show fit
|
0.12 |
ENST00000296679.9
ENST00000507029.5
|
WDR41
|
WD repeat domain 41
|
|
chr18_-_76495191
Show fit
|
0.09 |
ENST00000443185.7
|
ZNF516
|
zinc finger protein 516
|
|
chr12_+_62260338
Show fit
|
0.09 |
ENST00000353364.7
ENST00000549523.5
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr7_+_39950187
Show fit
|
0.08 |
ENST00000181839.10
|
CDK13
|
cyclin dependent kinase 13
|
|
chr4_+_39698109
Show fit
|
0.07 |
ENST00000510934.5
ENST00000261427.10
|
UBE2K
|
ubiquitin conjugating enzyme E2 K
|
|
chr12_+_104064520
Show fit
|
0.07 |
ENST00000229330.9
|
HCFC2
|
host cell factor C2
|
|
chr6_+_87155537
Show fit
|
0.06 |
ENST00000369577.8
ENST00000518845.1
ENST00000339907.8
ENST00000496806.2
|
ZNF292
|
zinc finger protein 292
|
|
chr18_+_65751000
Show fit
|
0.06 |
ENST00000397968.4
|
CDH7
|
cadherin 7
|
|
chr17_-_5486157
Show fit
|
0.06 |
ENST00000572834.5
ENST00000158771.9
ENST00000570848.5
ENST00000571971.1
|
DERL2
|
derlin 2
|
|
chr2_+_60756226
Show fit
|
0.05 |
ENST00000238714.8
|
PAPOLG
|
poly(A) polymerase gamma
|
|
chr13_-_49792675
Show fit
|
0.05 |
ENST00000261667.8
|
KPNA3
|
karyopherin subunit alpha 3
|
|
chr5_-_161546708
Show fit
|
0.05 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2
|
|
chr12_+_2052977
Show fit
|
0.05 |
ENST00000399634.6
ENST00000406454.8
ENST00000327702.12
ENST00000347598.9
ENST00000399603.6
ENST00000399641.6
ENST00000399655.6
ENST00000335762.10
ENST00000682835.1
|
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C
|
|
chr11_-_59668981
Show fit
|
0.05 |
ENST00000300146.10
|
PATL1
|
PAT1 homolog 1, processing body mRNA decay factor
|
|
chr17_+_29390326
Show fit
|
0.05 |
ENST00000261716.8
|
TAOK1
|
TAO kinase 1
|
|
chr17_-_28951285
Show fit
|
0.05 |
ENST00000577226.5
|
PHF12
|
PHD finger protein 12
|
|
chr14_-_21269451
Show fit
|
0.05 |
ENST00000336053.10
|
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C
|
|
chr2_+_105744876
Show fit
|
0.04 |
ENST00000233154.9
ENST00000451463.6
|
NCK2
|
NCK adaptor protein 2
|
|
chr10_+_27504234
Show fit
|
0.04 |
ENST00000621805.5
ENST00000535776.6
ENST00000684501.1
ENST00000683797.1
ENST00000682389.1
ENST00000356940.11
ENST00000683755.1
ENST00000682963.1
|
RAB18
|
RAB18, member RAS oncogene family
|
|
chr9_+_92325910
Show fit
|
0.04 |
ENST00000375587.8
ENST00000618653.1
|
CENPP
|
centromere protein P
|
|
chr4_+_61201223
Show fit
|
0.04 |
ENST00000512091.6
|
ADGRL3
|
adhesion G protein-coupled receptor L3
|
|
chr6_+_45328203
Show fit
|
0.04 |
ENST00000371432.7
ENST00000647337.2
ENST00000371438.5
|
RUNX2
|
RUNX family transcription factor 2
|
|
chr21_-_15064934
Show fit
|
0.04 |
ENST00000400199.5
ENST00000400202.5
ENST00000318948.7
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr5_+_83471736
Show fit
|
0.04 |
ENST00000265077.8
|
VCAN
|
versican
|
|
chr12_+_123584523
Show fit
|
0.04 |
ENST00000438031.2
ENST00000262225.8
|
TMED2
|
transmembrane p24 trafficking protein 2
|
|
chr1_+_193121950
Show fit
|
0.04 |
ENST00000367435.5
|
CDC73
|
cell division cycle 73
|
|
chr11_-_57324907
Show fit
|
0.04 |
ENST00000358252.8
|
TNKS1BP1
|
tankyrase 1 binding protein 1
|
|
chr14_+_32939243
Show fit
|
0.04 |
ENST00000346562.6
ENST00000548645.5
ENST00000356141.8
ENST00000357798.9
|
NPAS3
|
neuronal PAS domain protein 3
|
|
chr8_+_60516947
Show fit
|
0.03 |
ENST00000262646.12
|
RAB2A
|
RAB2A, member RAS oncogene family
|
|
chr3_-_177196451
Show fit
|
0.03 |
ENST00000430069.5
ENST00000630796.2
ENST00000428970.5
|
TBL1XR1
|
TBL1X receptor 1
|
|
chr5_-_132737518
Show fit
|
0.03 |
ENST00000403231.6
ENST00000378735.5
ENST00000618515.4
ENST00000378746.8
|
KIF3A
|
kinesin family member 3A
|
|
chr19_-_32675139
Show fit
|
0.03 |
ENST00000586693.7
ENST00000587352.5
ENST00000306065.9
ENST00000586463.5
|
ANKRD27
|
ankyrin repeat domain 27
|
|
chr5_-_159099909
Show fit
|
0.03 |
ENST00000313708.11
|
EBF1
|
EBF transcription factor 1
|
|
chr19_-_38849923
Show fit
|
0.03 |
ENST00000601813.1
ENST00000221419.10
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L
|
|
chr17_+_59565598
Show fit
|
0.03 |
ENST00000251241.9
ENST00000425628.7
ENST00000584385.5
ENST00000580030.1
|
DHX40
|
DEAH-box helicase 40
|
|
chr10_+_110919595
Show fit
|
0.03 |
ENST00000369452.9
|
SHOC2
|
SHOC2 leucine rich repeat scaffold protein
|
|
chr6_+_150865665
Show fit
|
0.03 |
ENST00000611279.4
ENST00000367321.8
ENST00000367307.8
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like
|
|
chr8_-_81112055
Show fit
|
0.03 |
ENST00000220597.4
|
PAG1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1
|
|
chr12_+_12717359
Show fit
|
0.03 |
ENST00000614874.2
ENST00000228872.9
|
CDKN1B
|
cyclin dependent kinase inhibitor 1B
|
|
chr6_-_145814744
Show fit
|
0.03 |
ENST00000237281.5
|
FBXO30
|
F-box protein 30
|
|
chr17_-_48604959
Show fit
|
0.03 |
ENST00000225648.4
ENST00000484302.3
|
HOXB6
|
homeobox B6
|
|
chr15_+_58987652
Show fit
|
0.03 |
ENST00000348370.9
ENST00000559160.1
|
RNF111
|
ring finger protein 111
|
|
chr10_-_92243246
Show fit
|
0.03 |
ENST00000412050.8
ENST00000614585.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr20_+_36306325
Show fit
|
0.03 |
ENST00000373913.7
ENST00000339266.10
|
DLGAP4
|
DLG associated protein 4
|
|
chr18_+_46334007
Show fit
|
0.03 |
ENST00000269439.12
ENST00000590330.1
|
RNF165
|
ring finger protein 165
|
|
chr2_-_201451446
Show fit
|
0.03 |
ENST00000332624.8
ENST00000430254.1
|
TRAK2
|
trafficking kinesin protein 2
|
|
chr2_-_152099023
Show fit
|
0.02 |
ENST00000201943.10
ENST00000427385.6
ENST00000539935.7
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4
|
|
chr4_+_41935423
Show fit
|
0.02 |
ENST00000504986.6
|
TMEM33
|
transmembrane protein 33
|
|
chr19_-_5978078
Show fit
|
0.02 |
ENST00000592621.5
ENST00000034275.12
ENST00000591092.5
ENST00000591333.5
ENST00000590623.5
ENST00000439268.6
|
RANBP3
|
RAN binding protein 3
|
|
chr1_-_10796636
Show fit
|
0.02 |
ENST00000377022.8
ENST00000344008.5
|
CASZ1
|
castor zinc finger 1
|
|
chr17_+_31936993
Show fit
|
0.02 |
ENST00000322652.10
|
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit
|
|
chr4_+_7192519
Show fit
|
0.02 |
ENST00000507866.6
|
SORCS2
|
sortilin related VPS10 domain containing receptor 2
|
|
chr7_-_155812454
Show fit
|
0.02 |
ENST00000297261.7
|
SHH
|
sonic hedgehog signaling molecule
|
|
chr11_+_9384621
Show fit
|
0.02 |
ENST00000379719.8
ENST00000527431.1
ENST00000630083.1
|
IPO7
|
importin 7
|
|
chr1_-_217089627
Show fit
|
0.02 |
ENST00000361525.7
|
ESRRG
|
estrogen related receptor gamma
|
|
chr12_+_113221429
Show fit
|
0.02 |
ENST00000551096.5
ENST00000551099.5
ENST00000552897.5
ENST00000550785.5
ENST00000549279.1
ENST00000335509.11
|
TPCN1
|
two pore segment channel 1
|
|
chr12_-_94459854
Show fit
|
0.01 |
ENST00000397809.10
|
CEP83
|
centrosomal protein 83
|
|
chr22_+_38681941
Show fit
|
0.01 |
ENST00000216034.6
|
TOMM22
|
translocase of outer mitochondrial membrane 22
|
|
chr20_-_50153637
Show fit
|
0.01 |
ENST00000341698.2
ENST00000371652.9
ENST00000371650.9
|
PEDS1-UBE2V1
PEDS1
|
PEDS1-UBE2V1 readthrough
plasmanylethanolamine desaturase 1
|
|
chr20_-_62065834
Show fit
|
0.01 |
ENST00000252996.9
|
TAF4
|
TATA-box binding protein associated factor 4
|
|
chr14_-_69153106
Show fit
|
0.01 |
ENST00000341516.10
|
DCAF5
|
DDB1 and CUL4 associated factor 5
|
|
chr13_-_51453015
Show fit
|
0.01 |
ENST00000442263.4
ENST00000311234.9
|
INTS6
|
integrator complex subunit 6
|
|
chrX_+_12138426
Show fit
|
0.01 |
ENST00000380682.5
ENST00000675598.1
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr8_+_74824526
Show fit
|
0.01 |
ENST00000649643.1
ENST00000260113.7
|
PI15
|
peptidase inhibitor 15
|
|
chr5_+_144205250
Show fit
|
0.01 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16
|
|
chr3_+_31532901
Show fit
|
0.01 |
ENST00000295770.4
|
STT3B
|
STT3 oligosaccharyltransferase complex catalytic subunit B
|
|
chr12_-_100142830
Show fit
|
0.01 |
ENST00000356828.7
ENST00000279907.12
|
UHRF1BP1L
|
UHRF1 binding protein 1 like
|
|
chr1_-_204359885
Show fit
|
0.01 |
ENST00000414478.1
ENST00000272203.8
|
PLEKHA6
|
pleckstrin homology domain containing A6
|
|
chr4_-_170003738
Show fit
|
0.01 |
ENST00000502832.1
ENST00000393704.3
|
MFAP3L
|
microfibril associated protein 3 like
|
|
chr1_-_70205531
Show fit
|
0.01 |
ENST00000370952.4
|
LRRC40
|
leucine rich repeat containing 40
|
|
chr5_+_173888335
Show fit
|
0.01 |
ENST00000265085.10
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr1_-_114511160
Show fit
|
0.01 |
ENST00000369543.6
ENST00000358465.7
|
TRIM33
|
tripartite motif containing 33
|
|
chr17_+_50719565
Show fit
|
0.01 |
ENST00000625349.2
ENST00000393227.6
ENST00000240304.5
ENST00000505658.6
ENST00000505619.5
ENST00000510984.5
|
LUC7L3
|
LUC7 like 3 pre-mRNA splicing factor
|
|
chrX_+_41085436
Show fit
|
0.01 |
ENST00000324545.9
ENST00000378308.7
|
USP9X
|
ubiquitin specific peptidase 9 X-linked
|
|
chr4_-_48906788
Show fit
|
0.01 |
ENST00000273860.8
|
OCIAD2
|
OCIA domain containing 2
|
|
chr6_+_104957099
Show fit
|
0.01 |
ENST00000345080.5
|
LIN28B
|
lin-28 homolog B
|
|
chr8_-_8893548
Show fit
|
0.00 |
ENST00000276282.7
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr1_+_22563460
Show fit
|
0.00 |
ENST00000166244.8
ENST00000374644.8
|
EPHA8
|
EPH receptor A8
|
|
chr5_+_142108753
Show fit
|
0.00 |
ENST00000253814.6
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr1_+_36306359
Show fit
|
0.00 |
ENST00000453908.8
|
SH3D21
|
SH3 domain containing 21
|
|
chr20_-_44810539
Show fit
|
0.00 |
ENST00000372851.8
|
RIMS4
|
regulating synaptic membrane exocytosis 4
|
|
chr9_-_89178810
Show fit
|
0.00 |
ENST00000375835.9
|
SHC3
|
SHC adaptor protein 3
|
|
chr7_+_38977904
Show fit
|
0.00 |
ENST00000518318.7
ENST00000403058.6
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr3_+_62319037
Show fit
|
0.00 |
ENST00000494481.5
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr2_-_212538766
Show fit
|
0.00 |
ENST00000342788.9
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4
|
|
chrX_+_44873169
Show fit
|
0.00 |
ENST00000675577.1
ENST00000674867.1
ENST00000674586.1
ENST00000382899.9
ENST00000536777.6
ENST00000543216.6
ENST00000377967.9
ENST00000611820.5
|
KDM6A
|
lysine demethylase 6A
|
|
chr5_-_41510554
Show fit
|
0.00 |
ENST00000377801.8
|
PLCXD3
|
phosphatidylinositol specific phospholipase C X domain containing 3
|