Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
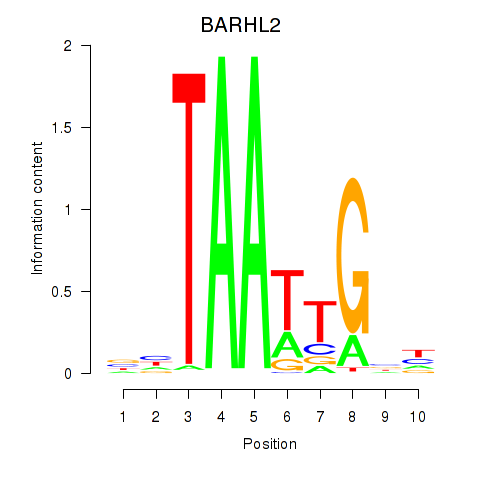
Results for BARHL2
Z-value: 1.27
Transcription factors associated with BARHL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARHL2
|
ENSG00000143032.8 | BARHL2 |
Activity profile of BARHL2 motif
Sorted Z-values of BARHL2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of BARHL2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_41613476 | 4.81 |
ENST00000508466.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr11_-_33892010 | 4.74 |
ENST00000257818.3
|
LMO2
|
LIM domain only 2 |
| chr1_+_84181630 | 4.33 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr4_+_155666718 | 4.26 |
ENST00000621234.4
ENST00000511108.5 |
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr6_-_75493629 | 3.92 |
ENST00000393004.6
|
FILIP1
|
filamin A interacting protein 1 |
| chr6_-_75493773 | 3.57 |
ENST00000237172.12
|
FILIP1
|
filamin A interacting protein 1 |
| chr2_-_207167220 | 3.15 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr2_-_187448244 | 3.00 |
ENST00000392370.8
ENST00000410068.5 ENST00000447403.5 ENST00000410102.5 |
CALCRL
|
calcitonin receptor like receptor |
| chr3_+_40100007 | 2.87 |
ENST00000539167.2
|
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr2_-_224402097 | 2.70 |
ENST00000409685.4
|
FAM124B
|
family with sequence similarity 124 member B |
| chr17_+_41226648 | 2.64 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2 |
| chr9_+_6215786 | 2.64 |
ENST00000417746.6
ENST00000682010.1 |
IL33
|
interleukin 33 |
| chr9_+_72577939 | 2.62 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr3_-_71305986 | 2.32 |
ENST00000647614.1
|
FOXP1
|
forkhead box P1 |
| chr17_+_68525795 | 2.27 |
ENST00000592800.5
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr2_-_187554351 | 2.21 |
ENST00000437725.5
ENST00000409676.5 ENST00000233156.9 ENST00000339091.8 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor |
| chr3_+_132597260 | 2.19 |
ENST00000249887.3
|
ACKR4
|
atypical chemokine receptor 4 |
| chr12_+_93572664 | 2.14 |
ENST00000551556.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr2_-_224401994 | 2.11 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124 member B |
| chr9_-_14180779 | 2.09 |
ENST00000380924.1
ENST00000543693.5 |
NFIB
|
nuclear factor I B |
| chr11_+_59787067 | 2.08 |
ENST00000528805.1
|
STX3
|
syntaxin 3 |
| chr4_+_143381939 | 2.07 |
ENST00000505913.5
|
GAB1
|
GRB2 associated binding protein 1 |
| chr1_+_84164370 | 2.03 |
ENST00000446538.5
ENST00000610703.4 ENST00000370682.7 ENST00000394838.6 ENST00000432111.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr16_-_58546702 | 1.97 |
ENST00000567133.1
|
CNOT1
|
CCR4-NOT transcription complex subunit 1 |
| chr19_+_44751251 | 1.91 |
ENST00000444487.1
|
BCL3
|
BCL3 transcription coactivator |
| chrX_-_45200895 | 1.89 |
ENST00000377934.4
|
DIPK2B
|
divergent protein kinase domain 2B |
| chr3_+_183253795 | 1.83 |
ENST00000460419.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chrX_-_117973717 | 1.78 |
ENST00000262820.7
|
KLHL13
|
kelch like family member 13 |
| chr12_-_64752871 | 1.76 |
ENST00000418919.6
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr9_+_2159672 | 1.75 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_-_22194709 | 1.72 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr4_-_25863537 | 1.68 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chrX_+_10158448 | 1.65 |
ENST00000380829.5
ENST00000421085.7 ENST00000674669.1 ENST00000454850.1 |
CLCN4
|
chloride voltage-gated channel 4 |
| chr20_+_43916142 | 1.65 |
ENST00000423191.6
ENST00000372999.5 |
TOX2
|
TOX high mobility group box family member 2 |
| chr12_-_91153149 | 1.62 |
ENST00000550758.1
|
DCN
|
decorin |
| chr4_+_133149278 | 1.61 |
ENST00000264360.7
|
PCDH10
|
protocadherin 10 |
| chrX_-_117973579 | 1.61 |
ENST00000371878.5
|
KLHL13
|
kelch like family member 13 |
| chr12_-_6124662 | 1.61 |
ENST00000261405.10
|
VWF
|
von Willebrand factor |
| chr18_+_44700796 | 1.58 |
ENST00000677130.1
|
SETBP1
|
SET binding protein 1 |
| chr1_+_47023659 | 1.56 |
ENST00000371901.4
|
CYP4X1
|
cytochrome P450 family 4 subfamily X member 1 |
| chr9_+_2159850 | 1.54 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_-_110179995 | 1.52 |
ENST00000392586.5
ENST00000419252.1 ENST00000359451.6 ENST00000392588.5 |
WASF1
|
WASP family member 1 |
| chr1_+_84301694 | 1.51 |
ENST00000394834.8
ENST00000370669.5 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr13_-_44474296 | 1.51 |
ENST00000611198.4
|
TSC22D1
|
TSC22 domain family member 1 |
| chr7_+_107583919 | 1.48 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr4_-_89835617 | 1.47 |
ENST00000611107.1
ENST00000345009.8 ENST00000505199.5 ENST00000502987.5 |
SNCA
|
synuclein alpha |
| chr8_+_11809135 | 1.47 |
ENST00000528643.5
ENST00000525777.5 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr6_+_29100609 | 1.47 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor family 2 subfamily J member 1 |
| chr6_-_75363003 | 1.46 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr12_-_7695752 | 1.45 |
ENST00000329913.4
|
GDF3
|
growth differentiation factor 3 |
| chr11_-_104898670 | 1.43 |
ENST00000422698.6
|
CASP12
|
caspase 12 (gene/pseudogene) |
| chr2_-_207166818 | 1.42 |
ENST00000423015.5
|
KLF7
|
Kruppel like factor 7 |
| chr5_-_111976925 | 1.40 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr9_-_90642855 | 1.39 |
ENST00000637905.1
|
DIRAS2
|
DIRAS family GTPase 2 |
| chr4_+_87006736 | 1.39 |
ENST00000544085.6
|
AFF1
|
AF4/FMR2 family member 1 |
| chr4_-_69860138 | 1.38 |
ENST00000226444.4
|
SULT1E1
|
sulfotransferase family 1E member 1 |
| chrX_+_136169833 | 1.34 |
ENST00000628032.2
|
FHL1
|
four and a half LIM domains 1 |
| chr1_-_16978276 | 1.32 |
ENST00000375534.7
|
MFAP2
|
microfibril associated protein 2 |
| chr17_+_58692563 | 1.29 |
ENST00000461271.5
ENST00000583539.5 ENST00000337432.9 ENST00000421782.3 |
RAD51C
|
RAD51 paralog C |
| chrX_-_45200828 | 1.28 |
ENST00000398000.7
|
DIPK2B
|
divergent protein kinase domain 2B |
| chr14_+_21990357 | 1.28 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chrX_+_136169664 | 1.25 |
ENST00000456445.5
|
FHL1
|
four and a half LIM domains 1 |
| chr16_+_30395400 | 1.25 |
ENST00000320159.2
ENST00000613509.2 |
ZNF48
|
zinc finger protein 48 |
| chr13_-_52011337 | 1.21 |
ENST00000400366.6
ENST00000400370.8 ENST00000634844.1 ENST00000673772.1 ENST00000418097.7 ENST00000242839.10 ENST00000344297.9 ENST00000448424.7 |
ATP7B
|
ATPase copper transporting beta |
| chr4_-_88284747 | 1.20 |
ENST00000514204.1
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr2_+_108621260 | 1.19 |
ENST00000409441.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr19_+_49513353 | 1.19 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr6_-_83709382 | 1.19 |
ENST00000520302.5
ENST00000520213.5 ENST00000439399.6 |
SNAP91
|
synaptosome associated protein 91 |
| chr6_+_143677935 | 1.17 |
ENST00000440869.6
ENST00000367582.7 ENST00000451827.6 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr1_+_196819731 | 1.16 |
ENST00000320493.10
ENST00000367424.4 |
CFHR1
|
complement factor H related 1 |
| chr11_-_122116215 | 1.14 |
ENST00000560104.2
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr7_+_100119607 | 1.13 |
ENST00000262932.5
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr16_+_86566821 | 1.12 |
ENST00000649859.1
|
FOXC2
|
forkhead box C2 |
| chr1_+_153774210 | 1.11 |
ENST00000271857.6
|
SLC27A3
|
solute carrier family 27 member 3 |
| chr1_-_94121105 | 1.10 |
ENST00000649773.1
ENST00000370225.4 |
ABCA4
|
ATP binding cassette subfamily A member 4 |
| chr3_+_183253230 | 1.10 |
ENST00000326505.4
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chrX_-_18672101 | 1.09 |
ENST00000379984.4
|
RS1
|
retinoschisin 1 |
| chrX_+_136169624 | 1.08 |
ENST00000394153.6
|
FHL1
|
four and a half LIM domains 1 |
| chr11_+_118533032 | 1.08 |
ENST00000526853.1
|
TMEM25
|
transmembrane protein 25 |
| chr6_-_154430495 | 1.07 |
ENST00000424998.3
|
CNKSR3
|
CNKSR family member 3 |
| chr2_+_33134579 | 1.07 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr18_-_12656716 | 1.07 |
ENST00000462226.1
ENST00000497844.6 ENST00000309836.9 ENST00000453447.6 |
SPIRE1
|
spire type actin nucleation factor 1 |
| chr3_+_37975773 | 1.07 |
ENST00000436654.1
|
CTDSPL
|
CTD small phosphatase like |
| chr15_+_24954912 | 1.07 |
ENST00000584968.5
ENST00000346403.10 ENST00000554227.6 ENST00000390687.9 ENST00000579070.5 ENST00000577565.1 ENST00000577949.5 ENST00000338327.4 |
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N SNRPN upstream reading frame |
| chr5_+_36166556 | 1.06 |
ENST00000677886.1
|
SKP2
|
S-phase kinase associated protein 2 |
| chr4_-_162163955 | 1.06 |
ENST00000379164.8
|
FSTL5
|
follistatin like 5 |
| chr2_-_182522703 | 1.05 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A |
| chr9_+_113463697 | 1.05 |
ENST00000317613.10
|
RGS3
|
regulator of G protein signaling 3 |
| chr2_+_172860038 | 1.05 |
ENST00000538974.5
ENST00000540783.5 |
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr9_+_12775012 | 1.03 |
ENST00000319264.4
|
LURAP1L
|
leucine rich adaptor protein 1 like |
| chr1_-_110391041 | 1.03 |
ENST00000369781.8
ENST00000437429.6 ENST00000541986.5 |
SLC16A4
|
solute carrier family 16 member 4 |
| chrX_+_87517784 | 1.02 |
ENST00000373119.9
ENST00000373114.4 |
KLHL4
|
kelch like family member 4 |
| chr2_-_182242031 | 0.99 |
ENST00000358139.6
|
PDE1A
|
phosphodiesterase 1A |
| chr4_-_162163989 | 0.99 |
ENST00000306100.10
ENST00000427802.2 |
FSTL5
|
follistatin like 5 |
| chr1_+_77281963 | 0.99 |
ENST00000354567.7
|
AK5
|
adenylate kinase 5 |
| chrX_-_16712865 | 0.99 |
ENST00000380241.7
|
CTPS2
|
CTP synthase 2 |
| chr8_+_22567038 | 0.97 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr15_-_37101205 | 0.96 |
ENST00000338564.9
ENST00000558313.5 ENST00000340545.9 |
MEIS2
|
Meis homeobox 2 |
| chr12_-_10807286 | 0.96 |
ENST00000240615.3
|
TAS2R8
|
taste 2 receptor member 8 |
| chr3_+_141426108 | 0.95 |
ENST00000441582.2
ENST00000510726.1 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr11_-_101129706 | 0.92 |
ENST00000534013.5
|
PGR
|
progesterone receptor |
| chr1_-_201171545 | 0.89 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9 |
| chr11_+_34642366 | 0.89 |
ENST00000532302.1
|
EHF
|
ETS homologous factor |
| chr12_-_71157992 | 0.89 |
ENST00000247829.8
|
TSPAN8
|
tetraspanin 8 |
| chr5_-_88877967 | 0.88 |
ENST00000508610.5
ENST00000636294.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr11_-_125111708 | 0.87 |
ENST00000531909.5
ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr7_-_129952901 | 0.87 |
ENST00000472396.5
ENST00000355621.8 |
UBE2H
|
ubiquitin conjugating enzyme E2 H |
| chr5_+_73813518 | 0.86 |
ENST00000296799.8
|
ARHGEF28
|
Rho guanine nucleotide exchange factor 28 |
| chrX_+_101078861 | 0.85 |
ENST00000372930.5
|
TMEM35A
|
transmembrane protein 35A |
| chr11_+_102112445 | 0.85 |
ENST00000524575.5
|
YAP1
|
Yes1 associated transcriptional regulator |
| chr16_+_8712943 | 0.83 |
ENST00000561870.5
ENST00000396600.6 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr14_-_50561119 | 0.83 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr7_+_12687625 | 0.82 |
ENST00000651779.1
ENST00000404894.1 |
ARL4A
|
ADP ribosylation factor like GTPase 4A |
| chr17_+_69502397 | 0.81 |
ENST00000613873.4
ENST00000589647.5 |
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_+_155308748 | 0.80 |
ENST00000611010.4
ENST00000447866.5 ENST00000368356.9 ENST00000467076.5 ENST00000491013.5 ENST00000356657.10 |
FDPS
|
farnesyl diphosphate synthase |
| chr12_-_95116967 | 0.79 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr4_+_122339221 | 0.79 |
ENST00000442707.1
|
KIAA1109
|
KIAA1109 |
| chr5_-_134174765 | 0.78 |
ENST00000520417.1
|
SKP1
|
S-phase kinase associated protein 1 |
| chr16_-_15094008 | 0.78 |
ENST00000327307.11
|
RRN3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr4_-_137532452 | 0.78 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr4_-_48780242 | 0.78 |
ENST00000507711.5
ENST00000358350.9 |
FRYL
|
FRY like transcription coactivator |
| chr4_+_89901979 | 0.77 |
ENST00000508372.1
|
MMRN1
|
multimerin 1 |
| chr3_+_111911604 | 0.77 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology like domain family B member 2 |
| chr6_+_143608170 | 0.77 |
ENST00000427704.6
ENST00000305766.10 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chrX_+_71301742 | 0.77 |
ENST00000373829.8
ENST00000538820.1 |
ITGB1BP2
|
integrin subunit beta 1 binding protein 2 |
| chr6_-_52994248 | 0.76 |
ENST00000457564.1
ENST00000370960.5 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr13_-_44436801 | 0.76 |
ENST00000261489.6
|
TSC22D1
|
TSC22 domain family member 1 |
| chr5_+_62578810 | 0.76 |
ENST00000334994.6
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr20_-_37261808 | 0.76 |
ENST00000373614.7
|
GHRH
|
growth hormone releasing hormone |
| chr10_-_89251768 | 0.76 |
ENST00000336233.10
|
LIPA
|
lipase A, lysosomal acid type |
| chrX_+_55452119 | 0.76 |
ENST00000342972.3
|
MAGEH1
|
MAGE family member H1 |
| chr3_-_165837412 | 0.75 |
ENST00000479451.5
ENST00000488954.1 ENST00000264381.8 |
BCHE
|
butyrylcholinesterase |
| chr7_+_150368189 | 0.75 |
ENST00000519397.1
ENST00000479668.5 |
REPIN1
|
replication initiator 1 |
| chr4_-_109801978 | 0.75 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr7_+_80133830 | 0.75 |
ENST00000648098.1
ENST00000648476.1 ENST00000648412.1 ENST00000648953.1 ENST00000648306.1 ENST00000648832.1 ENST00000648877.1 ENST00000442586.2 ENST00000649487.1 ENST00000649267.1 |
GNAI1
|
G protein subunit alpha i1 |
| chrX_-_16712572 | 0.74 |
ENST00000359276.9
|
CTPS2
|
CTP synthase 2 |
| chr4_+_107824555 | 0.73 |
ENST00000394684.8
|
SGMS2
|
sphingomyelin synthase 2 |
| chr10_+_21524627 | 0.73 |
ENST00000651097.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr17_-_445939 | 0.72 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr11_-_104164361 | 0.72 |
ENST00000302251.9
|
PDGFD
|
platelet derived growth factor D |
| chr15_+_67521104 | 0.71 |
ENST00000342683.6
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chrX_+_38801417 | 0.71 |
ENST00000614558.2
ENST00000457894.5 |
MID1IP1
|
MID1 interacting protein 1 |
| chr7_-_120858066 | 0.71 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr7_+_116222804 | 0.70 |
ENST00000393481.6
|
TES
|
testin LIM domain protein |
| chr7_-_56034133 | 0.70 |
ENST00000421626.5
|
PSPH
|
phosphoserine phosphatase |
| chr9_-_5833014 | 0.69 |
ENST00000339450.10
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr11_-_118264445 | 0.69 |
ENST00000438295.2
|
MPZL2
|
myelin protein zero like 2 |
| chr13_+_35476740 | 0.69 |
ENST00000537702.5
|
NBEA
|
neurobeachin |
| chr11_+_102317492 | 0.69 |
ENST00000673846.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr16_+_56961942 | 0.68 |
ENST00000200676.8
ENST00000566128.1 |
CETP
|
cholesteryl ester transfer protein |
| chr10_+_70404129 | 0.67 |
ENST00000373218.5
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr5_-_179617581 | 0.67 |
ENST00000523449.5
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr21_-_38661694 | 0.67 |
ENST00000417133.6
ENST00000398910.5 ENST00000442448.5 ENST00000429727.6 |
ERG
|
ETS transcription factor ERG |
| chr11_+_114439424 | 0.67 |
ENST00000544196.5
ENST00000265881.10 ENST00000539754.5 ENST00000539275.5 |
REXO2
|
RNA exonuclease 2 |
| chrX_+_38801451 | 0.67 |
ENST00000378474.3
ENST00000336949.7 |
MID1IP1
|
MID1 interacting protein 1 |
| chr1_+_84164684 | 0.66 |
ENST00000370680.5
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr9_+_96928310 | 0.63 |
ENST00000354649.7
|
NUTM2G
|
NUT family member 2G |
| chr1_+_210328244 | 0.63 |
ENST00000541565.5
ENST00000413764.6 |
HHAT
|
hedgehog acyltransferase |
| chr17_-_42136431 | 0.63 |
ENST00000552162.5
ENST00000550504.5 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr12_-_54258275 | 0.62 |
ENST00000552562.1
|
CBX5
|
chromobox 5 |
| chr6_+_28281555 | 0.62 |
ENST00000259883.3
ENST00000682144.1 |
PGBD1
|
piggyBac transposable element derived 1 |
| chr15_+_99105071 | 0.62 |
ENST00000328642.11
ENST00000594047.2 ENST00000336292.11 |
SYNM
|
synemin |
| chr14_-_22957128 | 0.62 |
ENST00000342454.12
ENST00000555986.5 ENST00000554516.5 ENST00000347758.6 ENST00000206474.11 ENST00000555040.5 |
HAUS4
|
HAUS augmin like complex subunit 4 |
| chr12_-_91179472 | 0.61 |
ENST00000550099.5
ENST00000546391.5 |
DCN
|
decorin |
| chr14_-_22957100 | 0.61 |
ENST00000555367.5
|
HAUS4
|
HAUS augmin like complex subunit 4 |
| chr9_+_131289685 | 0.61 |
ENST00000372264.4
|
PLPP7
|
phospholipid phosphatase 7 (inactive) |
| chr14_-_22957061 | 0.60 |
ENST00000557591.5
ENST00000541587.6 ENST00000490506.5 ENST00000554406.1 |
HAUS4
|
HAUS augmin like complex subunit 4 |
| chrX_-_71255060 | 0.60 |
ENST00000373988.5
ENST00000373998.5 |
ZMYM3
|
zinc finger MYM-type containing 3 |
| chr15_+_58410543 | 0.59 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr20_+_11917859 | 0.59 |
ENST00000618296.4
ENST00000378226.7 |
BTBD3
|
BTB domain containing 3 |
| chr12_-_89352395 | 0.59 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr4_+_169620509 | 0.58 |
ENST00000347613.8
|
CLCN3
|
chloride voltage-gated channel 3 |
| chr9_-_21202205 | 0.58 |
ENST00000239347.3
|
IFNA7
|
interferon alpha 7 |
| chr8_-_85341659 | 0.58 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr5_+_151259793 | 0.57 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr3_+_52794768 | 0.56 |
ENST00000621946.4
ENST00000416872.6 ENST00000449956.2 |
ITIH3
|
inter-alpha-trypsin inhibitor heavy chain 3 |
| chrX_+_136169891 | 0.56 |
ENST00000449474.5
|
FHL1
|
four and a half LIM domains 1 |
| chr17_+_48723179 | 0.56 |
ENST00000422730.4
|
PRAC2
|
PRAC2 small nuclear protein |
| chr6_+_106098933 | 0.56 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1 |
| chr8_+_28891304 | 0.56 |
ENST00000355231.9
|
HMBOX1
|
homeobox containing 1 |
| chr5_+_119452467 | 0.56 |
ENST00000509514.6
ENST00000682996.1 |
HSD17B4
|
hydroxysteroid 17-beta dehydrogenase 4 |
| chr5_+_136160986 | 0.56 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr1_+_196943738 | 0.55 |
ENST00000367415.8
ENST00000367421.5 ENST00000649283.1 ENST00000476712.6 ENST00000496448.6 ENST00000473386.1 ENST00000649960.1 |
CFHR2
|
complement factor H related 2 |
| chr1_-_243163310 | 0.55 |
ENST00000492145.1
ENST00000490813.5 ENST00000464936.5 |
CEP170
|
centrosomal protein 170 |
| chr1_-_205680486 | 0.55 |
ENST00000367145.4
|
SLC45A3
|
solute carrier family 45 member 3 |
| chr7_+_101817601 | 0.54 |
ENST00000292535.12
ENST00000546411.7 ENST00000549414.6 ENST00000550008.6 ENST00000556210.1 |
CUX1
|
cut like homeobox 1 |
| chr1_+_32072919 | 0.54 |
ENST00000438825.5
ENST00000336294.10 |
TMEM39B
|
transmembrane protein 39B |
| chr2_+_218270392 | 0.53 |
ENST00000248451.7
ENST00000273077.9 |
PNKD
|
PNKD metallo-beta-lactamase domain containing |
| chr11_-_78341876 | 0.53 |
ENST00000340149.6
|
GAB2
|
GRB2 associated binding protein 2 |
| chr2_+_188292814 | 0.53 |
ENST00000409580.5
ENST00000409637.7 |
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr13_+_48256214 | 0.53 |
ENST00000650237.1
|
ITM2B
|
integral membrane protein 2B |
| chr1_+_248508073 | 0.53 |
ENST00000641804.1
|
OR2G6
|
olfactory receptor family 2 subfamily G member 6 |
| chr19_+_44905785 | 0.53 |
ENST00000446996.5
ENST00000252486.9 ENST00000434152.5 |
APOE
|
apolipoprotein E |
| chr8_-_33567118 | 0.53 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122 |
| chr17_-_49646581 | 0.52 |
ENST00000510476.5
ENST00000503676.5 |
SPOP
|
speckle type BTB/POZ protein |
| chr2_-_202870761 | 0.52 |
ENST00000420558.5
ENST00000418208.5 |
ICA1L
|
islet cell autoantigen 1 like |
| chr10_-_93482326 | 0.52 |
ENST00000359263.9
|
MYOF
|
myoferlin |
| chr9_+_12693327 | 0.52 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chr6_-_99425269 | 0.51 |
ENST00000647811.1
ENST00000481229.2 ENST00000369239.10 ENST00000681611.1 ENST00000681615.1 ENST00000438806.5 |
PNISR
|
PNN interacting serine and arginine rich protein |
| chr20_+_4686320 | 0.51 |
ENST00000430350.2
|
PRNP
|
prion protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.0 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.6 | 1.9 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.5 | 1.4 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.4 | 2.6 | GO:0061517 | microglial cell activation involved in immune response(GO:0002282) macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.4 | 1.7 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.4 | 1.2 | GO:0060003 | copper ion export(GO:0060003) |
| 0.4 | 1.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.4 | 1.1 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.4 | 1.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.3 | 2.8 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 0.8 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.3 | 1.3 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.3 | 0.8 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.3 | 0.8 | GO:1990654 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.2 | 1.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.2 | 1.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.5 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 2.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 1.5 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.2 | 0.7 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.2 | 1.4 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.2 | 0.7 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.2 | 3.0 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 0.4 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.2 | 2.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 2.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 0.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 1.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.2 | 2.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 2.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 1.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.2 | 1.5 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 | 0.5 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.2 | 0.9 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.2 | 0.5 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 1.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 0.5 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.2 | 4.7 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.2 | 1.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 2.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 | 0.7 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.2 | 0.8 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 0.8 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.2 | 0.6 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.2 | 2.6 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.0 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.5 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.4 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.1 | 1.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.4 | GO:0034092 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.1 | 1.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 1.8 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 0.5 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.3 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.7 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.9 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) regulation of synaptic activity(GO:0060025) |
| 0.1 | 0.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.4 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.3 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.1 | 0.3 | GO:0061011 | hepatic duct development(GO:0061011) hepatoblast differentiation(GO:0061017) |
| 0.1 | 2.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.6 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 2.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.4 | GO:0030821 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) |
| 0.1 | 0.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 1.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.3 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.2 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 0.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 1.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.9 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.3 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.2 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 1.4 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 0.2 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 | 0.2 | GO:0051257 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 1.9 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.3 | GO:0033084 | immature T cell proliferation in thymus(GO:0033080) regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.2 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.1 | 0.1 | GO:0060516 | primary prostatic bud elongation(GO:0060516) |
| 0.1 | 0.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.8 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 1.2 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.7 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.9 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 1.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.5 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 1.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.4 | GO:0071673 | positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.0 | 0.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.6 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.0 | 0.5 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 1.1 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 1.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.6 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 1.3 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 3.1 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 1.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 3.1 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.9 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 1.4 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.4 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.4 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.5 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.0 | 0.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.2 | GO:0043353 | slow-twitch skeletal muscle fiber contraction(GO:0031444) enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.9 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.0 | 0.5 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 1.3 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.8 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.3 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.4 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.9 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 1.0 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.2 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 | 0.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.0 | 0.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.6 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.2 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0097242 | beta-amyloid clearance(GO:0097242) |
| 0.0 | 0.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 3.3 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 2.8 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.0 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0051354 | negative regulation of oxidoreductase activity(GO:0051354) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 2.9 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.0 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.0 | GO:0048769 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) sarcomerogenesis(GO:0048769) |
| 0.0 | 1.0 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.4 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 2.4 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.1 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.6 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.9 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.0 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:1903027 | regulation of opsonization(GO:1903027) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.3 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.5 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.4 | 9.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 2.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.3 | 0.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.3 | 1.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 0.8 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.2 | 1.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.7 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 2.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 2.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 1.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.8 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 0.8 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 2.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.4 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.5 | GO:0072557 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 3.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 3.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 2.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.8 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.2 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 0.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.1 | 4.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.6 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.0 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0071756 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 2.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.4 | GO:0034358 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 3.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.7 | 3.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.4 | 7.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 1.7 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.4 | 1.3 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.4 | 1.2 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.4 | 1.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.4 | 1.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 1.1 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.3 | 2.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.3 | 1.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 2.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 0.8 | GO:0030290 | sphingolipid activator protein activity(GO:0030290) beta-N-acetylgalactosaminidase activity(GO:0032428) |
| 0.3 | 0.8 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.2 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 1.5 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 1.5 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.2 | 0.7 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.2 | 1.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 1.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 2.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 0.8 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.2 | 0.7 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.5 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 0.7 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.2 | 0.8 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 1.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.4 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 0.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.5 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 2.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.8 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 1.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 2.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.5 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.5 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 0.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 2.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.2 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.5 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.1 | 4.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.3 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.6 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.2 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.1 | 1.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.4 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 1.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 2.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 1.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.7 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 1.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 2.5 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 2.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 9.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 1.4 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 2.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 2.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 6.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 0.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 1.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 2.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.7 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 0.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.8 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.9 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.8 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 1.5 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 2.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |