Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for BHLHE22_BHLHA15_BHLHE23
Z-value: 0.31
Transcription factors associated with BHLHE22_BHLHA15_BHLHE23
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
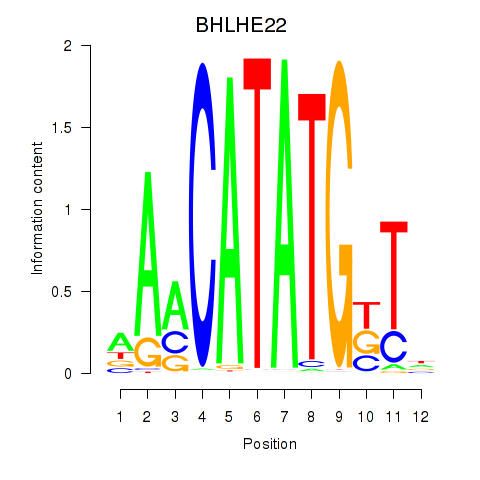
BHLHE22
|
ENSG00000180828.3 | BHLHE22 |
|
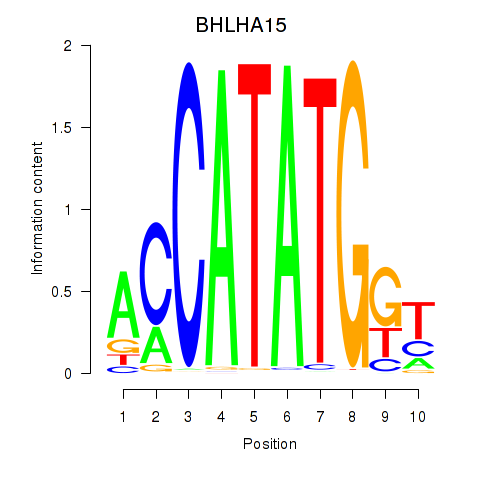
BHLHA15
|
ENSG00000180535.4 | BHLHA15 |
|
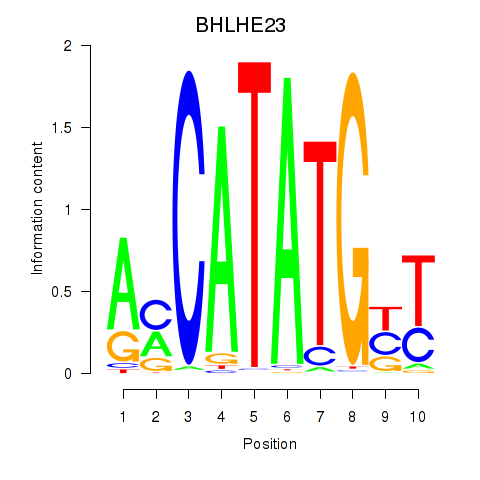
BHLHE23
|
ENSG00000125533.6 | BHLHE23 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE22 | hg38_v1_chr8_+_64580357_64580374 | 0.37 | 7.1e-02 | Click! |
| BHLHA15 | hg38_v1_chr7_+_98211431_98211474 | -0.36 | 7.4e-02 | Click! |
| BHLHE23 | hg38_v1_chr20_-_63006961_63007035 | -0.02 | 9.2e-01 | Click! |
Activity profile of BHLHE22_BHLHA15_BHLHE23 motif
Sorted Z-values of BHLHE22_BHLHA15_BHLHE23 motif
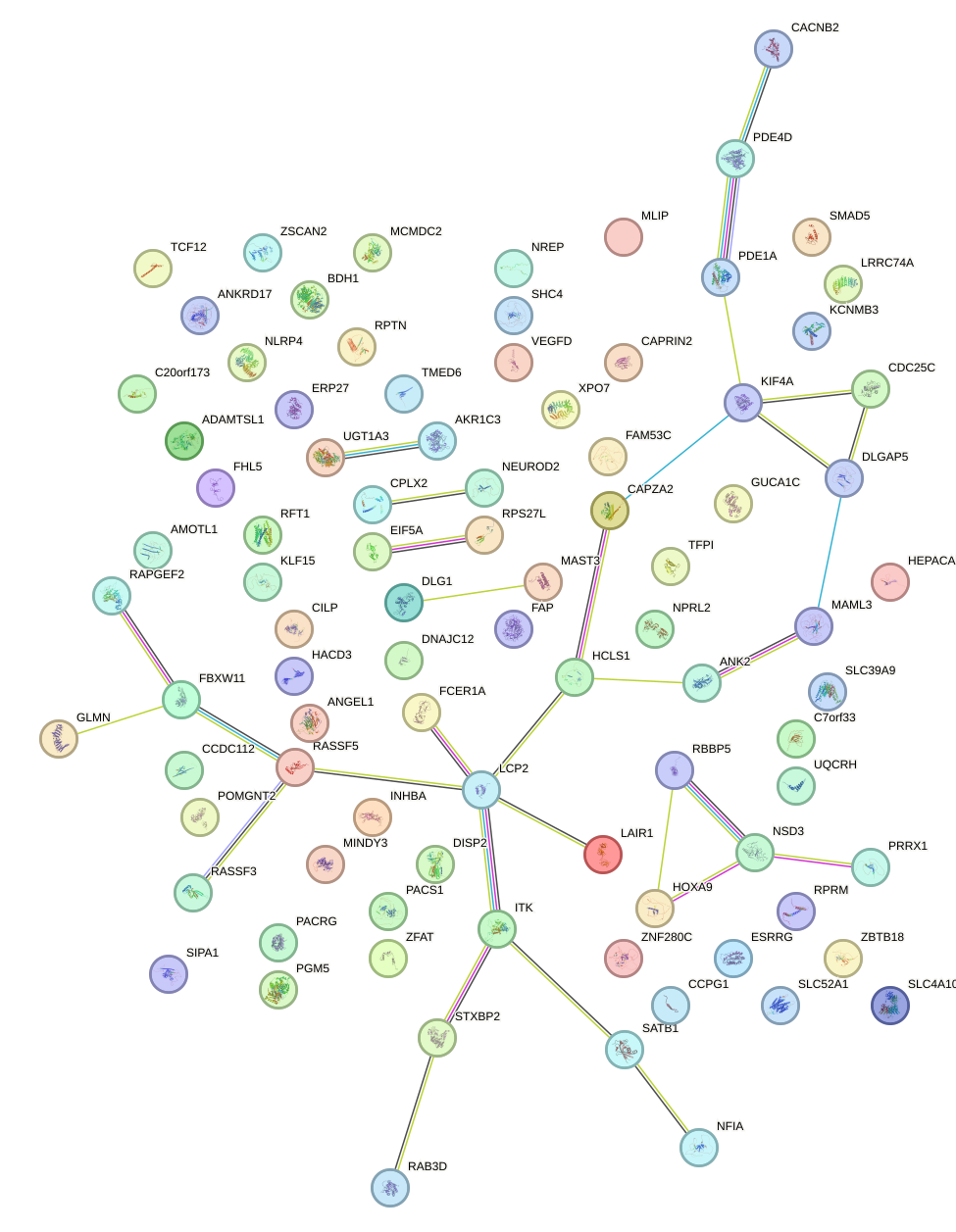
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE22_BHLHA15_BHLHE23
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_22207502 | 0.32 |
ENST00000390461.2
|
TRAV34
|
T cell receptor alpha variable 34 |
| chr16_-_69351778 | 0.28 |
ENST00000288025.4
|
TMED6
|
transmembrane p24 trafficking protein 6 |
| chr19_+_55857437 | 0.26 |
ENST00000587891.5
|
NLRP4
|
NLR family pyrin domain containing 4 |
| chr19_-_11339573 | 0.25 |
ENST00000222120.8
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr7_-_27165517 | 0.24 |
ENST00000396345.1
ENST00000343483.7 |
HOXA9
|
homeobox A9 |
| chr1_+_244051275 | 0.24 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr3_-_121660892 | 0.23 |
ENST00000428394.6
ENST00000314583.8 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr10_+_5094405 | 0.22 |
ENST00000380554.5
|
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr5_-_111976925 | 0.22 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr5_-_59430600 | 0.21 |
ENST00000636120.1
|
PDE4D
|
phosphodiesterase 4D |
| chr14_-_106875069 | 0.17 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr15_-_65211463 | 0.17 |
ENST00000261883.6
|
CILP
|
cartilage intermediate layer protein |
| chr5_+_175872741 | 0.16 |
ENST00000502265.5
|
CPLX2
|
complexin 2 |
| chr7_+_148590760 | 0.14 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr3_-_18438767 | 0.13 |
ENST00000454909.6
|
SATB1
|
SATB homeobox 1 |
| chr8_+_21966215 | 0.12 |
ENST00000433566.8
|
XPO7
|
exportin 7 |
| chr9_+_18474165 | 0.12 |
ENST00000380566.8
|
ADAMTSL1
|
ADAMTS like 1 |
| chr12_-_102478539 | 0.12 |
ENST00000424202.6
|
IGF1
|
insulin like growth factor 1 |
| chr15_+_65550819 | 0.12 |
ENST00000569894.5
|
HACD3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr1_+_60865259 | 0.12 |
ENST00000371191.5
|
NFIA
|
nuclear factor I A |
| chr7_+_116862552 | 0.12 |
ENST00000361183.8
ENST00000639546.1 ENST00000490693.5 |
CAPZA2
|
capping actin protein of muscle Z-line subunit alpha 2 |
| chr11_+_65638085 | 0.12 |
ENST00000534313.6
ENST00000533361.1 ENST00000526137.1 |
SIPA1
|
signal-induced proliferation-associated 1 |
| chr3_-_126357399 | 0.12 |
ENST00000296233.4
|
KLF15
|
Kruppel like factor 15 |
| chr15_-_55408018 | 0.11 |
ENST00000569205.5
|
CCPG1
|
cell cycle progression 1 |
| chr4_+_159267737 | 0.11 |
ENST00000264431.8
|
RAPGEF2
|
Rap guanine nucleotide exchange factor 2 |
| chr15_+_40358207 | 0.11 |
ENST00000267889.5
|
DISP2
|
dispatched RND transporter family member 2 |
| chr14_-_76826229 | 0.11 |
ENST00000557497.1
|
ANGEL1
|
angel homolog 1 |
| chr14_+_76826372 | 0.10 |
ENST00000393774.7
ENST00000555189.1 |
LRRC74A
|
leucine rich repeat containing 74A |
| chr6_+_162727129 | 0.10 |
ENST00000337019.7
ENST00000366889.6 |
PACRG
|
parkin coregulated |
| chr19_-_54364983 | 0.10 |
ENST00000434277.6
|
LAIR1
|
leukocyte associated immunoglobulin like receptor 1 |
| chr12_+_64610458 | 0.09 |
ENST00000542104.6
|
RASSF3
|
Ras association domain family member 3 |
| chr5_+_136160986 | 0.09 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr2_-_187513641 | 0.08 |
ENST00000392365.5
ENST00000435414.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr8_-_38382146 | 0.08 |
ENST00000534155.1
ENST00000433384.6 ENST00000317025.13 ENST00000316985.7 |
NSD3
|
nuclear receptor binding SET domain protein 3 |
| chrX_-_15384402 | 0.08 |
ENST00000297904.4
|
VEGFD
|
vascular endothelial growth factor D |
| chr5_-_138338325 | 0.08 |
ENST00000510119.1
ENST00000513970.5 |
CDC25C
|
cell division cycle 25C |
| chr7_-_41703062 | 0.08 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr17_-_49209367 | 0.07 |
ENST00000300406.6
ENST00000511277.5 ENST00000511673.1 |
GNGT2
|
G protein subunit gamma transducin 2 |
| chr5_-_170297746 | 0.07 |
ENST00000046794.10
|
LCP2
|
lymphocyte cytosolic protein 2 |
| chr3_-_197573323 | 0.07 |
ENST00000358186.6
ENST00000431056.5 |
BDH1
|
3-hydroxybutyrate dehydrogenase 1 |
| chr1_-_152159227 | 0.07 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr8_-_134510182 | 0.07 |
ENST00000521673.5
|
ZFAT
|
zinc finger and AT-hook domain containing |
| chr2_-_153478753 | 0.07 |
ENST00000325926.4
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator homolog |
| chr19_+_18097763 | 0.07 |
ENST00000262811.10
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr3_+_46883337 | 0.07 |
ENST00000313049.9
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr17_+_7308339 | 0.06 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr4_-_140154176 | 0.06 |
ENST00000509479.6
|
MAML3
|
mastermind like transcriptional coactivator 3 |
| chr17_+_7307961 | 0.06 |
ENST00000419711.6
ENST00000571955.5 ENST00000573714.5 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr12_-_14938508 | 0.06 |
ENST00000266397.7
|
ERP27
|
endoplasmic reticulum protein 27 |
| chr14_-_106025628 | 0.06 |
ENST00000631943.1
|
IGHV7-4-1
|
immunoglobulin heavy variable 7-4-1 |
| chr6_+_54083423 | 0.06 |
ENST00000460844.6
ENST00000370876.6 |
MLIP
|
muscular LMNA interacting protein |
| chr3_-_179266971 | 0.06 |
ENST00000349697.2
ENST00000497599.5 |
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr9_+_2158487 | 0.05 |
ENST00000634706.1
ENST00000634338.1 ENST00000635688.1 ENST00000634435.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr9_+_2158239 | 0.05 |
ENST00000635133.1
ENST00000634931.1 ENST00000423555.6 ENST00000382185.6 ENST00000302401.8 ENST00000382183.6 ENST00000417599.6 ENST00000382186.6 ENST00000635530.1 ENST00000635388.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_-_182522703 | 0.05 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A |
| chr3_-_197184131 | 0.05 |
ENST00000452595.5
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr3_-_197183849 | 0.05 |
ENST00000443183.5
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr15_-_48963912 | 0.05 |
ENST00000332408.9
|
SHC4
|
SHC adaptor protein 4 |
| chr3_-_43105939 | 0.05 |
ENST00000441964.1
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chrX_+_70290077 | 0.05 |
ENST00000374403.4
|
KIF4A
|
kinesin family member 4A |
| chr1_+_248508073 | 0.05 |
ENST00000641804.1
|
OR2G6
|
olfactory receptor family 2 subfamily G member 6 |
| chr20_-_35529618 | 0.05 |
ENST00000246199.5
ENST00000424444.1 ENST00000374345.8 ENST00000444723.3 |
C20orf173
|
chromosome 20 open reading frame 173 |
| chr2_-_162243375 | 0.05 |
ENST00000188790.9
ENST00000443424.5 |
FAP
|
fibroblast activation protein alpha |
| chr9_+_18474100 | 0.04 |
ENST00000327883.11
ENST00000431052.6 ENST00000380570.8 ENST00000380548.9 |
ADAMTSL1
|
ADAMTS like 1 |
| chr3_+_113211459 | 0.04 |
ENST00000495514.5
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr3_-_197183806 | 0.04 |
ENST00000671246.1
ENST00000660553.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr3_+_98149326 | 0.04 |
ENST00000437310.1
|
OR5H14
|
olfactory receptor family 5 subfamily H member 14 |
| chr1_+_170663917 | 0.04 |
ENST00000497230.2
|
PRRX1
|
paired related homeobox 1 |
| chr3_-_108953762 | 0.04 |
ENST00000393963.7
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr3_-_197183963 | 0.04 |
ENST00000653795.1
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr2_+_233729042 | 0.04 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chr7_-_93226449 | 0.04 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr1_-_205121986 | 0.03 |
ENST00000367164.1
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr3_-_108953870 | 0.03 |
ENST00000261047.8
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr4_+_113292838 | 0.03 |
ENST00000672411.1
ENST00000673231.1 |
ANK2
|
ankyrin 2 |
| chr15_-_63156774 | 0.03 |
ENST00000462430.5
|
RPS27L
|
ribosomal protein S27 like |
| chr6_+_96562548 | 0.03 |
ENST00000541107.5
ENST00000326771.2 |
FHL5
|
four and a half LIM domains 5 |
| chr4_+_113292925 | 0.03 |
ENST00000673353.1
ENST00000505342.6 ENST00000672915.1 ENST00000509550.5 |
ANK2
|
ankyrin 2 |
| chr12_-_30735014 | 0.03 |
ENST00000433722.6
|
CAPRIN2
|
caprin family member 2 |
| chr3_-_50350710 | 0.03 |
ENST00000232501.8
|
NPRL2
|
NPR2 like, GATOR1 complex subunit |
| chr3_-_53130405 | 0.03 |
ENST00000467048.1
ENST00000296292.8 ENST00000394738.7 |
RFT1
|
RFT1 homolog |
| chr12_+_50400809 | 0.03 |
ENST00000293618.12
ENST00000429001.7 ENST00000398473.7 ENST00000548174.5 ENST00000548697.5 ENST00000548993.5 ENST00000614335.4 ENST00000522085.5 ENST00000615080.4 ENST00000518444.5 ENST00000551886.5 ENST00000523389.5 ENST00000518561.5 ENST00000347328.9 ENST00000550260.1 |
LARP4
|
La ribonucleoprotein 4 |
| chr17_-_39607876 | 0.03 |
ENST00000302584.5
|
NEUROD2
|
neuronal differentiation 2 |
| chr3_+_113532508 | 0.03 |
ENST00000264852.9
|
SIDT1
|
SID1 transmembrane family member 1 |
| chr11_+_66241227 | 0.03 |
ENST00000531597.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr8_+_66870749 | 0.02 |
ENST00000396592.7
ENST00000422365.7 ENST00000492775.5 |
MCMDC2
|
minichromosome maintenance domain containing 2 |
| chr5_+_157180816 | 0.02 |
ENST00000422843.8
|
ITK
|
IL2 inducible T cell kinase |
| chr1_+_159302321 | 0.02 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE receptor Ia |
| chr1_+_46303646 | 0.02 |
ENST00000311672.10
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein |
| chr5_+_138338256 | 0.02 |
ENST00000513056.5
ENST00000239906.10 ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53 member C |
| chr4_-_73223082 | 0.02 |
ENST00000509867.6
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr2_-_182522556 | 0.02 |
ENST00000435564.5
|
PDE1A
|
phosphodiesterase 1A |
| chr4_-_158173042 | 0.02 |
ENST00000592057.1
ENST00000393807.9 |
GASK1B
|
golgi associated kinase 1B |
| chr9_+_68356603 | 0.02 |
ENST00000396396.6
|
PGM5
|
phosphoglucomutase 5 |
| chrX_-_130268883 | 0.02 |
ENST00000447817.1
ENST00000370978.9 |
ZNF280C
|
zinc finger protein 280C |
| chr16_+_21704963 | 0.02 |
ENST00000388957.3
|
OTOA
|
otoancorin |
| chr11_+_94768331 | 0.02 |
ENST00000317829.12
ENST00000433060.3 |
AMOTL1
|
angiomotin like 1 |
| chr5_-_115296610 | 0.02 |
ENST00000379611.10
ENST00000506442.5 |
CCDC112
|
coiled-coil domain containing 112 |
| chr10_-_67838019 | 0.02 |
ENST00000483798.6
|
DNAJC12
|
DnaJ heat shock protein family (Hsp40) member C12 |
| chr4_+_2812259 | 0.02 |
ENST00000502260.5
ENST00000435136.8 |
SH3BP2
|
SH3 domain binding protein 2 |
| chr1_-_217076889 | 0.02 |
ENST00000493748.5
ENST00000463665.5 |
ESRRG
|
estrogen related receptor gamma |
| chr10_+_18340821 | 0.02 |
ENST00000612743.1
ENST00000612134.4 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr15_+_84600986 | 0.01 |
ENST00000540936.1
ENST00000448803.6 ENST00000546275.1 ENST00000546148.6 ENST00000442073.3 ENST00000334141.7 ENST00000358472.3 ENST00000502939.2 ENST00000379358.7 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr14_+_69398683 | 0.01 |
ENST00000556605.5
ENST00000031146.8 ENST00000336643.10 |
SLC39A9
|
solute carrier family 39 member 9 |
| chr7_-_113118544 | 0.01 |
ENST00000397764.8
ENST00000441359.1 |
SMIM30
|
small integral membrane protein 30 |
| chr10_-_15860450 | 0.01 |
ENST00000277632.8
|
MINDY3
|
MINDY lysine 48 deubiquitinase 3 |
| chr2_+_161624335 | 0.01 |
ENST00000375514.9
ENST00000415876.6 |
SLC4A10
|
solute carrier family 4 member 10 |
| chr3_+_113211539 | 0.01 |
ENST00000682979.1
ENST00000485230.5 |
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr10_+_18340699 | 0.01 |
ENST00000377329.10
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr5_-_172006567 | 0.01 |
ENST00000517395.6
ENST00000265094.9 ENST00000393802.6 |
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr2_+_161624406 | 0.01 |
ENST00000446997.6
ENST00000272716.9 |
SLC4A10
|
solute carrier family 4 member 10 |
| chr15_+_57219411 | 0.01 |
ENST00000543579.5
ENST00000537840.5 ENST00000343827.7 |
TCF12
|
transcription factor 12 |
| chr14_-_50830641 | 0.01 |
ENST00000453196.6
ENST00000496749.1 |
NIN
|
ninein |
| chr4_-_158173004 | 0.01 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B |
| chr20_+_43565115 | 0.01 |
ENST00000423407.7
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr3_+_98168700 | 0.01 |
ENST00000383696.4
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15 |
| chr2_-_32265732 | 0.01 |
ENST00000360906.9
ENST00000342905.10 |
NLRC4
|
NLR family CARD domain containing 4 |
| chr8_+_67064316 | 0.01 |
ENST00000675306.2
ENST00000678017.1 ENST00000262210.11 ENST00000675869.1 ENST00000677009.1 ENST00000676847.1 ENST00000676471.1 ENST00000678542.1 ENST00000677619.1 ENST00000676605.1 ENST00000678553.1 ENST00000674993.1 ENST00000678318.1 ENST00000676573.1 ENST00000676317.1 ENST00000677592.1 ENST00000679226.1 ENST00000675955.1 ENST00000676882.1 ENST00000678616.1 ENST00000678645.1 ENST00000678747.1 |
CSPP1
|
centrosome and spindle pole associated protein 1 |
| chr14_-_50831046 | 0.01 |
ENST00000476352.5
ENST00000324330.13 ENST00000382041.7 ENST00000453401.6 ENST00000530997.7 |
NIN
|
ninein |
| chr5_-_172006817 | 0.01 |
ENST00000296933.10
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr7_+_107891135 | 0.01 |
ENST00000639772.1
ENST00000440410.5 ENST00000437604.6 ENST00000205402.10 |
DLD
|
dihydrolipoamide dehydrogenase |
| chr3_+_137998735 | 0.01 |
ENST00000343735.8
|
CLDN18
|
claudin 18 |
| chr3_-_33659441 | 0.01 |
ENST00000650653.1
ENST00000480013.6 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chrX_+_108091752 | 0.01 |
ENST00000457035.5
ENST00000372232.8 |
ATG4A
|
autophagy related 4A cysteine peptidase |
| chr6_+_72366730 | 0.01 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chrX_+_108091665 | 0.00 |
ENST00000345734.7
|
ATG4A
|
autophagy related 4A cysteine peptidase |
| chr3_-_33659097 | 0.00 |
ENST00000461133.8
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr10_-_102418748 | 0.00 |
ENST00000020673.6
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr14_-_50830479 | 0.00 |
ENST00000382043.8
|
NIN
|
ninein |
| chr14_-_69398276 | 0.00 |
ENST00000557016.6
ENST00000555373.1 |
ERH
|
ERH mRNA splicing and mitosis factor |
| chr11_-_62591500 | 0.00 |
ENST00000476907.6
ENST00000278279.7 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr11_-_62591554 | 0.00 |
ENST00000494385.1
ENST00000308436.11 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chrX_+_108044967 | 0.00 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr10_+_134465 | 0.00 |
ENST00000439456.5
ENST00000397962.8 ENST00000309776.8 ENST00000397959.7 |
ZMYND11
|
zinc finger MYND-type containing 11 |
| chr10_-_77090722 | 0.00 |
ENST00000638531.1
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr10_-_30999469 | 0.00 |
ENST00000538351.6
|
ZNF438
|
zinc finger protein 438 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.0 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.0 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |