Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for CBFB
Z-value: 0.38
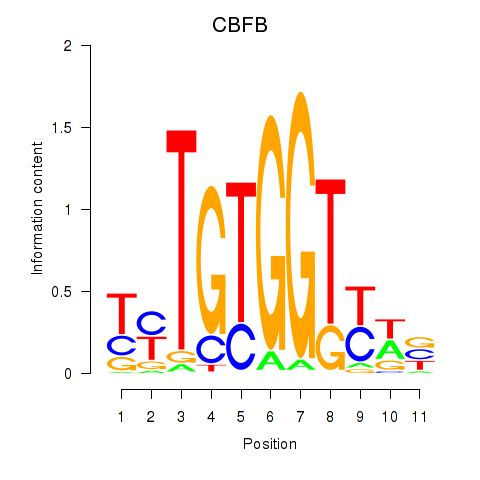
Transcription factors associated with CBFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CBFB
|
ENSG00000067955.15 | CBFB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CBFB | hg38_v1_chr16_+_67029133_67029185 | 0.20 | 3.4e-01 | Click! |
Activity profile of CBFB motif
Sorted Z-values of CBFB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CBFB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_132073782 | 1.47 |
ENST00000296871.4
|
CSF2
|
colony stimulating factor 2 |
| chr5_+_35856883 | 1.14 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr2_+_102418642 | 0.75 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr21_-_35049238 | 0.71 |
ENST00000416754.1
ENST00000437180.5 ENST00000455571.5 ENST00000675419.1 |
RUNX1
|
RUNX family transcription factor 1 |
| chr8_+_103372388 | 0.70 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr21_-_35049327 | 0.70 |
ENST00000300305.7
|
RUNX1
|
RUNX family transcription factor 1 |
| chrX_+_68829009 | 0.65 |
ENST00000204961.5
|
EFNB1
|
ephrin B1 |
| chr9_-_127778659 | 0.57 |
ENST00000314830.13
|
SH2D3C
|
SH2 domain containing 3C |
| chr3_-_190122317 | 0.40 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr8_-_133060347 | 0.36 |
ENST00000427060.6
|
SLA
|
Src like adaptor |
| chr12_+_14973020 | 0.34 |
ENST00000266395.3
|
PDE6H
|
phosphodiesterase 6H |
| chr18_-_28177934 | 0.29 |
ENST00000676445.1
|
CDH2
|
cadherin 2 |
| chr13_-_28100556 | 0.28 |
ENST00000241453.12
|
FLT3
|
fms related receptor tyrosine kinase 3 |
| chr11_-_58575846 | 0.23 |
ENST00000395074.7
|
LPXN
|
leupaxin |
| chr6_-_75202792 | 0.23 |
ENST00000416123.6
|
COL12A1
|
collagen type XII alpha 1 chain |
| chr14_-_60649449 | 0.23 |
ENST00000645694.3
|
SIX1
|
SIX homeobox 1 |
| chr8_-_95269190 | 0.22 |
ENST00000286688.6
|
C8orf37
|
chromosome 8 open reading frame 37 |
| chr17_+_66302606 | 0.22 |
ENST00000413366.8
|
PRKCA
|
protein kinase C alpha |
| chr7_+_120988683 | 0.21 |
ENST00000340646.9
ENST00000310396.10 |
CPED1
|
cadherin like and PC-esterase domain containing 1 |
| chr11_-_16356538 | 0.21 |
ENST00000683767.1
|
SOX6
|
SRY-box transcription factor 6 |
| chr17_+_34255274 | 0.20 |
ENST00000580907.5
ENST00000225831.4 |
CCL2
|
C-C motif chemokine ligand 2 |
| chr6_+_27957241 | 0.20 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor family 2 subfamily B member 6 |
| chr2_-_218985176 | 0.20 |
ENST00000295727.2
|
FEV
|
FEV transcription factor, ETS family member |
| chr1_+_209686173 | 0.18 |
ENST00000615289.4
ENST00000367028.6 ENST00000261465.5 |
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr4_+_73481737 | 0.17 |
ENST00000226355.5
|
AFM
|
afamin |
| chr4_+_87975667 | 0.16 |
ENST00000237623.11
ENST00000682655.1 ENST00000508233.6 ENST00000360804.4 ENST00000395080.8 |
SPP1
|
secreted phosphoprotein 1 |
| chr17_-_65560296 | 0.16 |
ENST00000585045.1
ENST00000611991.1 |
AXIN2
|
axin 2 |
| chr4_+_87975829 | 0.15 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1 |
| chr11_-_65117639 | 0.14 |
ENST00000528598.1
ENST00000310597.6 |
ZNHIT2
|
zinc finger HIT-type containing 2 |
| chr1_-_11796536 | 0.14 |
ENST00000641820.1
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr15_+_73684204 | 0.14 |
ENST00000537340.6
ENST00000318424.9 |
CD276
|
CD276 molecule |
| chr3_+_133124807 | 0.13 |
ENST00000508711.5
|
TMEM108
|
transmembrane protein 108 |
| chr2_-_127220293 | 0.13 |
ENST00000664447.2
ENST00000409327.2 |
CYP27C1
|
cytochrome P450 family 27 subfamily C member 1 |
| chr3_-_11568764 | 0.13 |
ENST00000424529.6
|
VGLL4
|
vestigial like family member 4 |
| chr19_-_13937991 | 0.13 |
ENST00000254320.7
ENST00000586075.1 |
PODNL1
|
podocan like 1 |
| chr3_+_157436842 | 0.12 |
ENST00000295927.4
|
PTX3
|
pentraxin 3 |
| chr1_-_184037695 | 0.11 |
ENST00000361927.9
ENST00000649786.1 |
COLGALT2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr15_+_49423233 | 0.11 |
ENST00000560270.1
ENST00000267843.9 ENST00000560979.1 |
FGF7
|
fibroblast growth factor 7 |
| chr9_+_15422704 | 0.11 |
ENST00000380821.7
ENST00000610884.4 ENST00000421710.5 |
SNAPC3
|
small nuclear RNA activating complex polypeptide 3 |
| chr19_-_13938371 | 0.11 |
ENST00000588872.3
ENST00000339560.10 |
PODNL1
|
podocan like 1 |
| chr14_-_59484317 | 0.11 |
ENST00000247194.9
|
L3HYPDH
|
trans-L-3-hydroxyproline dehydratase |
| chr22_+_31082860 | 0.10 |
ENST00000619644.4
|
SMTN
|
smoothelin |
| chr20_+_46008900 | 0.10 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 |
| chrX_-_49184789 | 0.10 |
ENST00000453382.5
ENST00000432913.5 |
PRICKLE3
|
prickle planar cell polarity protein 3 |
| chr4_-_155376797 | 0.10 |
ENST00000650955.1
ENST00000515654.5 |
MAP9
|
microtubule associated protein 9 |
| chr12_-_53200443 | 0.09 |
ENST00000550743.6
|
ITGB7
|
integrin subunit beta 7 |
| chr14_-_59484006 | 0.09 |
ENST00000481608.1
|
L3HYPDH
|
trans-L-3-hydroxyproline dehydratase |
| chr11_-_66336396 | 0.09 |
ENST00000627248.1
ENST00000311320.9 |
RIN1
|
Ras and Rab interactor 1 |
| chr1_+_17308194 | 0.06 |
ENST00000375453.5
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase 4 |
| chr12_+_80716906 | 0.06 |
ENST00000228644.4
|
MYF5
|
myogenic factor 5 |
| chr10_-_46023445 | 0.06 |
ENST00000585132.5
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr2_-_206159194 | 0.06 |
ENST00000455934.6
ENST00000449699.5 ENST00000454195.1 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr4_+_70226116 | 0.06 |
ENST00000317987.6
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr13_-_46182136 | 0.05 |
ENST00000323076.7
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr12_-_54634880 | 0.05 |
ENST00000547511.5
ENST00000257867.5 |
LACRT
|
lacritin |
| chr18_+_13611910 | 0.04 |
ENST00000590308.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr19_+_10430786 | 0.04 |
ENST00000293683.9
|
PDE4A
|
phosphodiesterase 4A |
| chr15_+_73683938 | 0.04 |
ENST00000567189.5
|
CD276
|
CD276 molecule |
| chr2_-_206159410 | 0.03 |
ENST00000457011.5
ENST00000440274.5 ENST00000432169.5 ENST00000233190.11 |
NDUFS1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr1_-_206772484 | 0.03 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr12_-_54384687 | 0.03 |
ENST00000550120.1
ENST00000547210.5 ENST00000394313.7 |
ZNF385A
|
zinc finger protein 385A |
| chr7_+_100969565 | 0.02 |
ENST00000536621.6
ENST00000379442.7 |
MUC12
|
mucin 12, cell surface associated |
| chr2_-_87021844 | 0.02 |
ENST00000355705.4
ENST00000409310.6 |
PLGLB1
|
plasminogen like B1 |
| chr5_+_132060648 | 0.02 |
ENST00000296870.3
|
IL3
|
interleukin 3 |
| chr6_+_52186373 | 0.02 |
ENST00000648244.1
|
IL17A
|
interleukin 17A |
| chr1_-_184754808 | 0.02 |
ENST00000318130.13
ENST00000367512.7 |
EDEM3
|
ER degradation enhancing alpha-mannosidase like protein 3 |
| chr5_-_39202991 | 0.02 |
ENST00000515010.5
|
FYB1
|
FYN binding protein 1 |
| chr20_+_56412112 | 0.00 |
ENST00000360314.7
|
CASS4
|
Cas scaffold protein family member 4 |
| chr1_+_34176900 | 0.00 |
ENST00000488417.2
|
C1orf94
|
chromosome 1 open reading frame 94 |
| chr16_+_53054973 | 0.00 |
ENST00000447540.6
ENST00000615216.4 ENST00000566029.5 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_-_120876356 | 0.00 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 1.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.7 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 1.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.3 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.2 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.2 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.2 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.2 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |