Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
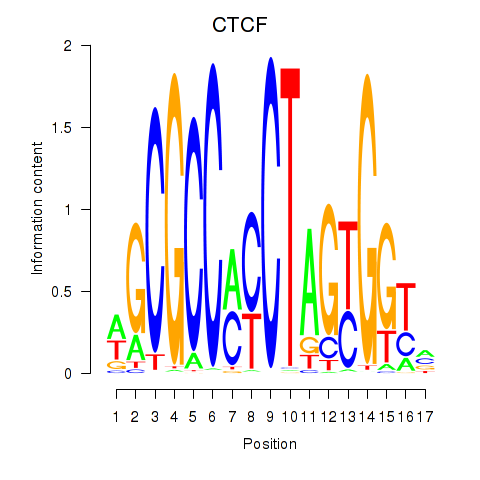
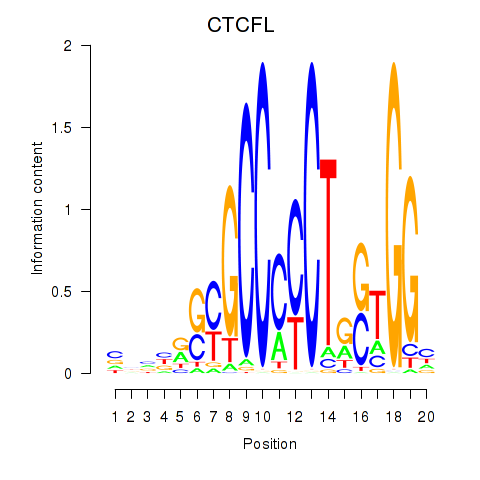
Results for CTCF_CTCFL
Z-value: 0.98
Transcription factors associated with CTCF_CTCFL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CTCF
|
ENSG00000102974.16 | CTCF |
|
CTCFL
|
ENSG00000124092.13 | CTCFL |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CTCFL | hg38_v1_chr20_-_57525097_57525108 | 0.21 | 3.0e-01 | Click! |
| CTCF | hg38_v1_chr16_+_67562514_67562594 | -0.21 | 3.1e-01 | Click! |
Activity profile of CTCF_CTCFL motif
Sorted Z-values of CTCF_CTCFL motif
Network of associatons between targets according to the STRING database.
First level regulatory network of CTCF_CTCFL
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_48524236 | 2.95 |
ENST00000374170.5
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr2_+_102355750 | 2.56 |
ENST00000233957.7
|
IL18R1
|
interleukin 18 receptor 1 |
| chr2_+_102355881 | 2.56 |
ENST00000409599.5
|
IL18R1
|
interleukin 18 receptor 1 |
| chr15_-_64046322 | 2.53 |
ENST00000457488.5
ENST00000612884.4 |
DAPK2
|
death associated protein kinase 2 |
| chr9_-_131740056 | 2.30 |
ENST00000372195.5
ENST00000683357.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor 1 |
| chr6_+_32854179 | 2.29 |
ENST00000374859.3
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr9_+_114155526 | 2.10 |
ENST00000356083.8
|
COL27A1
|
collagen type XXVII alpha 1 chain |
| chr6_-_32853618 | 2.01 |
ENST00000354258.5
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr6_-_32853813 | 1.98 |
ENST00000643049.2
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr11_-_72793636 | 1.97 |
ENST00000538536.5
ENST00000543304.5 ENST00000540587.1 |
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr16_-_2340703 | 1.77 |
ENST00000301732.10
ENST00000382381.7 |
ABCA3
|
ATP binding cassette subfamily A member 3 |
| chr11_-_57568276 | 1.71 |
ENST00000340573.8
|
UBE2L6
|
ubiquitin conjugating enzyme E2 L6 |
| chr10_+_13099440 | 1.69 |
ENST00000263036.9
|
OPTN
|
optineurin |
| chr14_+_24161257 | 1.65 |
ENST00000396864.8
ENST00000557894.5 ENST00000559284.5 ENST00000560275.5 |
IRF9
|
interferon regulatory factor 9 |
| chr1_-_184974477 | 1.61 |
ENST00000367511.4
|
NIBAN1
|
niban apoptosis regulator 1 |
| chr22_-_50526130 | 1.58 |
ENST00000535425.5
ENST00000638598.2 |
SCO2
|
synthesis of cytochrome C oxidase 2 |
| chr18_+_46174055 | 1.52 |
ENST00000615553.1
|
C18orf25
|
chromosome 18 open reading frame 25 |
| chr6_+_137867241 | 1.51 |
ENST00000612899.5
ENST00000420009.5 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr6_+_137867414 | 1.47 |
ENST00000237289.8
ENST00000433680.1 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr8_-_89984231 | 1.42 |
ENST00000517337.1
ENST00000409330.5 |
NBN
|
nibrin |
| chr10_+_102394488 | 1.41 |
ENST00000369966.8
|
NFKB2
|
nuclear factor kappa B subunit 2 |
| chr10_-_5978022 | 1.38 |
ENST00000525219.6
|
IL15RA
|
interleukin 15 receptor subunit alpha |
| chr15_+_74782069 | 1.37 |
ENST00000220003.14
ENST00000439220.6 |
CSK
|
C-terminal Src kinase |
| chr9_+_97412062 | 1.37 |
ENST00000355295.5
|
TDRD7
|
tudor domain containing 7 |
| chr22_-_50526337 | 1.36 |
ENST00000651490.1
ENST00000543927.6 |
TYMP
SCO2
|
thymidine phosphorylase synthesis of cytochrome C oxidase 2 |
| chr10_-_5977589 | 1.34 |
ENST00000620345.4
ENST00000397251.7 ENST00000397248.6 ENST00000622442.4 ENST00000620865.4 |
IL15RA
|
interleukin 15 receptor subunit alpha |
| chr3_-_190322434 | 1.32 |
ENST00000295522.4
|
CLDN1
|
claudin 1 |
| chr10_+_13099585 | 1.30 |
ENST00000378764.6
|
OPTN
|
optineurin |
| chr22_-_31107517 | 1.28 |
ENST00000400299.6
ENST00000611680.1 |
SELENOM
|
selenoprotein M |
| chr19_-_1848452 | 1.24 |
ENST00000170168.9
|
REXO1
|
RNA exonuclease 1 homolog |
| chr10_-_48604952 | 1.23 |
ENST00000417912.6
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr11_-_72794032 | 1.11 |
ENST00000334805.11
|
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr3_-_49929800 | 1.11 |
ENST00000455683.6
|
MON1A
|
MON1 homolog A, secretory trafficking associated |
| chr9_+_100427123 | 1.10 |
ENST00000395067.7
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3 |
| chr10_-_48605032 | 1.08 |
ENST00000249601.9
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr16_+_4624811 | 1.07 |
ENST00000415496.5
ENST00000262370.12 ENST00000587747.5 ENST00000399577.9 ENST00000588994.5 ENST00000586183.5 |
MGRN1
|
mahogunin ring finger 1 |
| chr2_+_233354474 | 1.05 |
ENST00000264057.7
ENST00000427930.5 ENST00000442524.4 |
DGKD
|
diacylglycerol kinase delta |
| chr8_-_89984609 | 1.03 |
ENST00000519426.5
ENST00000265433.8 |
NBN
|
nibrin |
| chr15_-_38564635 | 1.01 |
ENST00000450598.6
ENST00000559830.5 ENST00000558164.5 ENST00000539159.5 ENST00000310803.10 |
RASGRP1
|
RAS guanyl releasing protein 1 |
| chr22_+_31212207 | 0.99 |
ENST00000406516.5
ENST00000331728.9 |
LIMK2
|
LIM domain kinase 2 |
| chr22_-_50525548 | 0.97 |
ENST00000395693.8
ENST00000252785.3 |
SCO2
|
synthesis of cytochrome C oxidase 2 |
| chr10_-_78029487 | 0.91 |
ENST00000372371.8
|
POLR3A
|
RNA polymerase III subunit A |
| chr19_+_10871516 | 0.90 |
ENST00000327064.9
ENST00000588947.5 |
CARM1
|
coactivator associated arginine methyltransferase 1 |
| chr19_+_12838437 | 0.90 |
ENST00000251472.9
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr3_+_72888031 | 0.90 |
ENST00000389617.9
|
GXYLT2
|
glucoside xylosyltransferase 2 |
| chr5_+_173144442 | 0.88 |
ENST00000231668.13
ENST00000351486.10 ENST00000352523.10 ENST00000393770.4 |
BNIP1
|
BCL2 interacting protein 1 |
| chr11_-_65662780 | 0.87 |
ENST00000534283.1
ENST00000527749.5 ENST00000533187.5 ENST00000525693.5 ENST00000534558.5 ENST00000532879.5 ENST00000406246.8 ENST00000532999.5 |
RELA
|
RELA proto-oncogene, NF-kB subunit |
| chr19_+_10871673 | 0.86 |
ENST00000344150.8
|
CARM1
|
coactivator associated arginine methyltransferase 1 |
| chr19_+_40807112 | 0.86 |
ENST00000595621.1
ENST00000595051.1 |
EGLN2
|
egl-9 family hypoxia inducible factor 2 |
| chr10_-_15371225 | 0.85 |
ENST00000378116.9
|
FAM171A1
|
family with sequence similarity 171 member A1 |
| chr15_+_70892809 | 0.85 |
ENST00000260382.10
ENST00000560755.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr17_+_80101562 | 0.84 |
ENST00000302262.8
ENST00000577106.5 ENST00000390015.7 |
GAA
|
alpha glucosidase |
| chr19_-_48993300 | 0.83 |
ENST00000323798.8
ENST00000263276.6 |
GYS1
|
glycogen synthase 1 |
| chr20_-_31951970 | 0.82 |
ENST00000202017.6
|
PDRG1
|
p53 and DNA damage regulated 1 |
| chr1_+_21981099 | 0.82 |
ENST00000400277.2
|
CELA3B
|
chymotrypsin like elastase 3B |
| chr2_+_236569817 | 0.81 |
ENST00000272928.4
|
ACKR3
|
atypical chemokine receptor 3 |
| chr3_-_79019444 | 0.80 |
ENST00000618833.4
ENST00000436010.6 ENST00000618846.4 |
ROBO1
|
roundabout guidance receptor 1 |
| chr5_-_147453888 | 0.79 |
ENST00000398514.7
|
DPYSL3
|
dihydropyrimidinase like 3 |
| chr7_+_6104881 | 0.79 |
ENST00000306177.9
ENST00000465073.6 |
USP42
|
ubiquitin specific peptidase 42 |
| chr8_+_53851786 | 0.78 |
ENST00000297313.8
ENST00000344277.10 |
RGS20
|
regulator of G protein signaling 20 |
| chr6_-_128520358 | 0.78 |
ENST00000368215.7
ENST00000532331.5 ENST00000368213.9 ENST00000368207.7 ENST00000525459.1 ENST00000368226.9 ENST00000368210.7 |
PTPRK
|
protein tyrosine phosphatase receptor type K |
| chr9_+_100427254 | 0.77 |
ENST00000374885.5
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3 |
| chr17_-_75515509 | 0.75 |
ENST00000321617.8
|
CASKIN2
|
CASK interacting protein 2 |
| chr7_-_22500152 | 0.74 |
ENST00000406890.6
ENST00000678116.1 ENST00000424363.5 |
STEAP1B
|
STEAP family member 1B |
| chr6_+_125956696 | 0.74 |
ENST00000229633.7
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr20_+_37903104 | 0.71 |
ENST00000373459.4
ENST00000373461.9 ENST00000448944.1 |
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr16_+_2817180 | 0.70 |
ENST00000450020.7
|
PRSS21
|
serine protease 21 |
| chr10_-_94362925 | 0.69 |
ENST00000371361.3
|
NOC3L
|
NOC3 like DNA replication regulator |
| chrX_-_135871812 | 0.68 |
ENST00000620704.5
|
CT45A9
|
cancer/testis antigen family 45 member A9 |
| chr5_-_95961830 | 0.68 |
ENST00000513343.1
ENST00000237853.9 |
ELL2
|
elongation factor for RNA polymerase II 2 |
| chrX_-_135820012 | 0.67 |
ENST00000612907.2
|
CT45A2
|
cancer/testis antigen family 45 member A2 |
| chr21_+_6499203 | 0.67 |
ENST00000619537.5
|
CRYAA2
|
crystallin alpha A2 |
| chr5_+_321695 | 0.67 |
ENST00000684583.1
ENST00000514523.1 |
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr6_+_32844108 | 0.65 |
ENST00000458296.2
ENST00000413039.6 ENST00000412095.1 ENST00000395330.5 |
PSMB8-AS1
PSMB9
|
PSMB8 antisense RNA 1 (head to head) proteasome 20S subunit beta 9 |
| chr16_+_180435 | 0.65 |
ENST00000199708.3
|
HBQ1
|
hemoglobin subunit theta 1 |
| chrX_-_135785298 | 0.65 |
ENST00000487941.6
|
CT45A5
|
cancer/testis antigen family 45 member A5 |
| chr9_-_137046160 | 0.65 |
ENST00000371601.5
|
NPDC1
|
neural proliferation, differentiation and control 1 |
| chr6_+_125781108 | 0.65 |
ENST00000368357.7
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr14_+_21768482 | 0.65 |
ENST00000390428.3
|
TRAV6
|
T cell receptor alpha variable 6 |
| chr11_-_72793592 | 0.64 |
ENST00000536377.5
ENST00000359373.9 |
STARD10
ARAP1
|
StAR related lipid transfer domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr2_-_74392025 | 0.63 |
ENST00000440727.1
ENST00000409240.5 |
DCTN1
|
dynactin subunit 1 |
| chr18_+_12093839 | 0.62 |
ENST00000587848.3
|
ANKRD62
|
ankyrin repeat domain 62 |
| chr5_+_177087412 | 0.61 |
ENST00000513166.1
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr10_+_79347460 | 0.61 |
ENST00000225174.8
|
PPIF
|
peptidylprolyl isomerase F |
| chr12_+_48978313 | 0.61 |
ENST00000293549.4
|
WNT1
|
Wnt family member 1 |
| chr17_+_43483949 | 0.61 |
ENST00000540306.5
ENST00000262415.8 |
DHX8
|
DEAH-box helicase 8 |
| chr1_+_10210562 | 0.61 |
ENST00000377093.9
ENST00000676179.1 |
KIF1B
|
kinesin family member 1B |
| chrX_+_23783163 | 0.61 |
ENST00000379254.5
ENST00000379270.5 ENST00000683890.1 |
SAT1
ENSG00000288706.1
|
spermidine/spermine N1-acetyltransferase 1 novel protein |
| chr1_-_153549238 | 0.60 |
ENST00000368713.8
|
S100A3
|
S100 calcium binding protein A3 |
| chr19_-_38869921 | 0.60 |
ENST00000593809.1
ENST00000593424.5 |
RINL
|
Ras and Rab interactor like |
| chr3_-_187139477 | 0.60 |
ENST00000455270.5
ENST00000296277.9 |
RPL39L
|
ribosomal protein L39 like |
| chr17_-_1187294 | 0.59 |
ENST00000544583.6
|
ABR
|
ABR activator of RhoGEF and GTPase |
| chr22_-_18024513 | 0.59 |
ENST00000441493.7
|
MICAL3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr1_+_15153698 | 0.59 |
ENST00000400796.7
ENST00000376008.3 ENST00000434578.6 |
TMEM51
|
transmembrane protein 51 |
| chr17_-_8079903 | 0.58 |
ENST00000649809.1
|
ALOX12B
|
arachidonate 12-lipoxygenase, 12R type |
| chr22_+_31082860 | 0.58 |
ENST00000619644.4
|
SMTN
|
smoothelin |
| chr17_-_36534883 | 0.58 |
ENST00000620640.4
|
MYO19
|
myosin XIX |
| chr1_+_32741779 | 0.58 |
ENST00000401073.7
|
KIAA1522
|
KIAA1522 |
| chr16_-_69754913 | 0.58 |
ENST00000268802.10
|
NOB1
|
NIN1 (RPN12) binding protein 1 homolog |
| chr19_+_39412650 | 0.57 |
ENST00000425673.6
|
PLEKHG2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr14_+_104689588 | 0.56 |
ENST00000330634.11
ENST00000392634.9 ENST00000675482.1 ENST00000398337.8 |
INF2
|
inverted formin 2 |
| chr6_+_146543824 | 0.55 |
ENST00000367495.4
|
RAB32
|
RAB32, member RAS oncogene family |
| chr9_+_128203397 | 0.55 |
ENST00000628346.2
ENST00000486160.3 ENST00000627061.2 ENST00000627543.2 ENST00000634267.2 |
DNM1
|
dynamin 1 |
| chr7_-_6348906 | 0.55 |
ENST00000313324.9
ENST00000530143.1 |
FAM220A
|
family with sequence similarity 220 member A |
| chr1_+_10430720 | 0.55 |
ENST00000602296.6
|
CENPS-CORT
|
CENPS-CORT readthrough |
| chr18_-_50195138 | 0.54 |
ENST00000285039.12
|
MYO5B
|
myosin VB |
| chr6_-_32838727 | 0.54 |
ENST00000652259.1
ENST00000374897.4 ENST00000620123.4 ENST00000452392.2 |
TAP2
ENSG00000250264.1
|
transporter 2, ATP binding cassette subfamily B member novel protein, TAP2-HLA-DOB readthrough |
| chr9_+_128203371 | 0.54 |
ENST00000475805.5
ENST00000372923.8 ENST00000341179.11 ENST00000393594.7 |
DNM1
|
dynamin 1 |
| chr10_+_70815889 | 0.54 |
ENST00000373202.8
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chr2_-_89160329 | 0.53 |
ENST00000390256.2
|
IGKV6-21
|
immunoglobulin kappa variable 6-21 (non-functional) |
| chr20_+_49982969 | 0.53 |
ENST00000244050.3
|
SNAI1
|
snail family transcriptional repressor 1 |
| chr2_+_90021567 | 0.53 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr17_-_7614824 | 0.53 |
ENST00000571597.1
ENST00000250113.12 |
FXR2
|
FMR1 autosomal homolog 2 |
| chr11_+_2400488 | 0.52 |
ENST00000380996.9
ENST00000380992.5 ENST00000333256.11 ENST00000437110.5 ENST00000435795.5 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr16_+_2817230 | 0.52 |
ENST00000005995.8
ENST00000574813.5 |
PRSS21
|
serine protease 21 |
| chr7_-_33109363 | 0.52 |
ENST00000684207.1
ENST00000297157.8 |
RP9
|
RP9 pre-mRNA splicing factor |
| chr9_-_137578853 | 0.52 |
ENST00000277540.7
|
DPH7
|
diphthamide biosynthesis 7 |
| chr9_+_128420812 | 0.51 |
ENST00000372838.9
|
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr3_+_126983035 | 0.51 |
ENST00000393409.3
|
PLXNA1
|
plexin A1 |
| chr1_+_47438036 | 0.51 |
ENST00000334793.6
|
FOXD2
|
forkhead box D2 |
| chr8_+_41578176 | 0.51 |
ENST00000396987.7
ENST00000519853.5 |
GPAT4
|
glycerol-3-phosphate acyltransferase 4 |
| chr12_+_55681711 | 0.51 |
ENST00000394252.4
|
METTL7B
|
methyltransferase like 7B |
| chr5_-_141619049 | 0.51 |
ENST00000647433.1
ENST00000253811.10 ENST00000389057.9 ENST00000398557.8 |
DIAPH1
|
diaphanous related formin 1 |
| chr17_-_42681840 | 0.50 |
ENST00000332438.4
|
CCR10
|
C-C motif chemokine receptor 10 |
| chr2_+_90069662 | 0.50 |
ENST00000390271.2
|
IGKV6D-41
|
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr2_-_130181542 | 0.49 |
ENST00000441135.1
ENST00000680679.1 ENST00000680401.1 ENST00000351288.10 ENST00000680298.1 ENST00000431183.6 |
SMPD4
|
sphingomyelin phosphodiesterase 4 |
| chr12_+_48978453 | 0.49 |
ENST00000613114.4
|
WNT1
|
Wnt family member 1 |
| chr9_-_133149334 | 0.49 |
ENST00000393160.7
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chrX_-_135768191 | 0.49 |
ENST00000598716.3
|
CT45A3
|
cancer/testis antigen family 45 member A3 |
| chr16_+_84145256 | 0.49 |
ENST00000378553.10
|
DNAAF1
|
dynein axonemal assembly factor 1 |
| chr8_+_89757789 | 0.49 |
ENST00000220751.5
|
RIPK2
|
receptor interacting serine/threonine kinase 2 |
| chr5_-_141618957 | 0.49 |
ENST00000389054.8
|
DIAPH1
|
diaphanous related formin 1 |
| chr2_+_85753984 | 0.49 |
ENST00000306279.4
|
ATOH8
|
atonal bHLH transcription factor 8 |
| chr12_-_54391270 | 0.49 |
ENST00000352268.10
ENST00000549962.5 ENST00000338010.9 ENST00000550774.5 |
ZNF385A
|
zinc finger protein 385A |
| chr1_+_10430070 | 0.48 |
ENST00000400900.6
|
CENPS-CORT
|
CENPS-CORT readthrough |
| chr17_-_50397472 | 0.48 |
ENST00000576448.1
ENST00000225972.8 |
LRRC59
|
leucine rich repeat containing 59 |
| chr2_-_74391837 | 0.47 |
ENST00000417090.1
ENST00000409868.5 ENST00000680606.1 |
DCTN1
|
dynactin subunit 1 |
| chr20_+_62302896 | 0.46 |
ENST00000620230.4
ENST00000253003.7 |
ADRM1
|
adhesion regulating molecule 1 |
| chr11_-_44950839 | 0.46 |
ENST00000395648.7
ENST00000531928.6 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr12_-_53321243 | 0.46 |
ENST00000547757.2
|
AAAS
|
aladin WD repeat nucleoporin |
| chr3_+_101849505 | 0.46 |
ENST00000326151.9
ENST00000326172.9 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr10_+_6202866 | 0.46 |
ENST00000317350.8
ENST00000379785.5 ENST00000625260.2 ENST00000626882.2 ENST00000360521.7 ENST00000379775.9 ENST00000640683.1 |
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr11_+_66258467 | 0.46 |
ENST00000394066.6
|
KLC2
|
kinesin light chain 2 |
| chr10_+_102132994 | 0.45 |
ENST00000413464.6
ENST00000278070.7 |
PPRC1
|
PPARG related coactivator 1 |
| chr4_-_163332589 | 0.45 |
ENST00000296533.3
ENST00000509586.5 ENST00000504391.5 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chr22_-_42720813 | 0.45 |
ENST00000381278.4
|
A4GALT
|
alpha 1,4-galactosyltransferase (P blood group) |
| chr3_+_23203306 | 0.45 |
ENST00000396703.6
|
UBE2E2
|
ubiquitin conjugating enzyme E2 E2 |
| chr1_+_228165794 | 0.45 |
ENST00000366711.4
|
IBA57
|
iron-sulfur cluster assembly factor IBA57 |
| chr22_-_42720861 | 0.45 |
ENST00000642412.2
|
A4GALT
|
alpha 1,4-galactosyltransferase (P blood group) |
| chr19_-_3547306 | 0.45 |
ENST00000589063.5
ENST00000615073.4 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr15_-_77420135 | 0.45 |
ENST00000560626.6
|
PEAK1
|
pseudopodium enriched atypical kinase 1 |
| chr16_-_50681328 | 0.45 |
ENST00000300590.7
|
SNX20
|
sorting nexin 20 |
| chr1_+_32753991 | 0.44 |
ENST00000373481.7
|
KIAA1522
|
KIAA1522 |
| chr9_+_34646589 | 0.44 |
ENST00000450095.6
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chr3_-_47475811 | 0.44 |
ENST00000265565.10
ENST00000428413.5 |
SCAP
|
SREBF chaperone |
| chr9_+_34646654 | 0.43 |
ENST00000378842.8
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chr15_+_91853690 | 0.43 |
ENST00000318445.11
|
SLCO3A1
|
solute carrier organic anion transporter family member 3A1 |
| chr11_+_65014103 | 0.43 |
ENST00000246747.9
ENST00000529384.5 ENST00000533729.1 |
ARL2
|
ADP ribosylation factor like GTPase 2 |
| chr19_+_14072754 | 0.43 |
ENST00000587086.2
|
MISP3
|
MISP family member 3 |
| chr15_-_77420087 | 0.42 |
ENST00000564328.5
ENST00000682557.1 ENST00000558305.5 |
PEAK1
|
pseudopodium enriched atypical kinase 1 |
| chr9_-_83063159 | 0.42 |
ENST00000340717.4
|
RASEF
|
RAS and EF-hand domain containing |
| chr9_+_89605004 | 0.42 |
ENST00000252506.11
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA damage inducible gamma |
| chr9_+_97501622 | 0.42 |
ENST00000259365.9
|
TMOD1
|
tropomodulin 1 |
| chr19_+_41262656 | 0.42 |
ENST00000599719.5
ENST00000601309.5 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U like 1 |
| chr17_-_76726753 | 0.41 |
ENST00000617192.4
|
JMJD6
|
jumonji domain containing 6, arginine demethylase and lysine hydroxylase |
| chr17_-_19362732 | 0.40 |
ENST00000395616.7
|
B9D1
|
B9 domain containing 1 |
| chr1_+_205256189 | 0.40 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr11_+_64241600 | 0.40 |
ENST00000535135.7
ENST00000652094.1 |
FKBP2
|
FKBP prolyl isomerase 2 |
| chr12_-_53321544 | 0.40 |
ENST00000394384.7
ENST00000209873.9 |
AAAS
|
aladin WD repeat nucleoporin |
| chr5_+_35617951 | 0.40 |
ENST00000440995.6
|
SPEF2
|
sperm flagellar 2 |
| chr1_-_145859061 | 0.40 |
ENST00000393045.7
|
PIAS3
|
protein inhibitor of activated STAT 3 |
| chr6_-_33580229 | 0.39 |
ENST00000374467.4
ENST00000442998.6 ENST00000360661.9 |
BAK1
|
BCL2 antagonist/killer 1 |
| chr9_+_69035729 | 0.39 |
ENST00000642889.1
ENST00000646862.1 ENST00000484259.3 ENST00000396366.6 |
ENSG00000285130.2
FXN
|
novel protein frataxin |
| chr8_+_22165140 | 0.39 |
ENST00000397814.7
ENST00000354870.5 |
BMP1
|
bone morphogenetic protein 1 |
| chr1_-_145859043 | 0.39 |
ENST00000369298.5
|
PIAS3
|
protein inhibitor of activated STAT 3 |
| chr9_+_100427379 | 0.39 |
ENST00000622639.4
ENST00000374886.7 |
MSANTD3
|
Myb/SANT DNA binding domain containing 3 |
| chr7_+_149147422 | 0.39 |
ENST00000475153.6
|
ZNF398
|
zinc finger protein 398 |
| chr5_-_141618914 | 0.39 |
ENST00000518047.5
|
DIAPH1
|
diaphanous related formin 1 |
| chr1_+_16440700 | 0.38 |
ENST00000504551.6
ENST00000457722.6 ENST00000337132.10 ENST00000443980.6 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr3_-_51941874 | 0.38 |
ENST00000232888.7
|
RRP9
|
ribosomal RNA processing 9, U3 small nucleolar RNA binding protein |
| chr1_+_24319342 | 0.38 |
ENST00000361548.9
|
GRHL3
|
grainyhead like transcription factor 3 |
| chr7_-_4883683 | 0.38 |
ENST00000399583.4
|
RADIL
|
Rap associating with DIL domain |
| chr11_-_44950867 | 0.38 |
ENST00000528290.5
ENST00000525680.6 ENST00000530035.5 ENST00000527685.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr20_+_50190821 | 0.38 |
ENST00000303004.5
|
CEBPB
|
CCAAT enhancer binding protein beta |
| chr18_-_12884151 | 0.37 |
ENST00000591115.5
ENST00000309660.10 ENST00000353319.8 ENST00000327283.7 |
PTPN2
|
protein tyrosine phosphatase non-receptor type 2 |
| chr18_-_12883766 | 0.37 |
ENST00000645191.1
ENST00000646492.1 |
PTPN2
|
protein tyrosine phosphatase non-receptor type 2 |
| chr1_+_10430384 | 0.37 |
ENST00000470413.6
ENST00000602787.6 ENST00000309048.8 |
CENPS-CORT
CENPS
|
CENPS-CORT readthrough centromere protein S |
| chr11_-_66438788 | 0.37 |
ENST00000329819.4
ENST00000310999.11 ENST00000430466.6 |
MRPL11
|
mitochondrial ribosomal protein L11 |
| chr11_-_36510232 | 0.37 |
ENST00000348124.5
ENST00000526995.6 |
TRAF6
|
TNF receptor associated factor 6 |
| chr16_-_57536543 | 0.37 |
ENST00000258214.3
|
CCDC102A
|
coiled-coil domain containing 102A |
| chr14_+_51651858 | 0.37 |
ENST00000395718.6
|
FRMD6
|
FERM domain containing 6 |
| chr17_-_44376169 | 0.36 |
ENST00000587295.5
|
ITGA2B
|
integrin subunit alpha 2b |
| chr5_-_139482285 | 0.36 |
ENST00000652110.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1 |
| chr9_-_38069220 | 0.36 |
ENST00000377707.4
|
SHB
|
SH2 domain containing adaptor protein B |
| chr15_+_70853239 | 0.36 |
ENST00000544974.6
ENST00000558546.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr12_-_49707220 | 0.36 |
ENST00000550488.5
|
FMNL3
|
formin like 3 |
| chrX_-_64230600 | 0.36 |
ENST00000362002.3
|
ASB12
|
ankyrin repeat and SOCS box containing 12 |
| chr11_+_46347526 | 0.36 |
ENST00000456247.6
ENST00000421244.6 ENST00000318201.12 |
DGKZ
|
diacylglycerol kinase zeta |
| chr1_-_51345105 | 0.35 |
ENST00000413473.6
ENST00000401051.7 ENST00000527205.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr6_-_17987463 | 0.35 |
ENST00000378814.9
ENST00000636847.1 ENST00000378843.6 ENST00000378826.6 ENST00000502704.2 ENST00000259711.11 |
KIF13A
|
kinesin family member 13A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 1.0 | 3.0 | GO:0034148 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.6 | 5.1 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.6 | 1.8 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) histone H3-R2 methylation(GO:0034970) |
| 0.5 | 1.8 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.4 | 3.0 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.4 | 2.5 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.4 | 2.9 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.4 | 1.2 | GO:0061184 | positive regulation of dermatome development(GO:0061184) |
| 0.4 | 1.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.4 | 1.4 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.4 | 2.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.3 | 1.0 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.3 | 0.9 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.3 | 0.9 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.3 | 3.7 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.3 | 0.8 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.2 | 0.7 | GO:1902232 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) regulation of positive thymic T cell selection(GO:1902232) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.2 | 2.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 0.8 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of negative chemotaxis(GO:0050925) |
| 0.2 | 1.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.2 | 0.9 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.2 | 0.7 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.2 | 0.5 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.2 | 1.0 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.2 | 1.3 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.2 | 0.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.2 | 0.6 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.1 | 3.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.1 | GO:2000078 | positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 | 0.4 | GO:0018395 | peptidyl-lysine hydroxylation to 5-hydroxy-L-lysine(GO:0018395) histone arginine demethylation(GO:0070077) histone H3-R2 demethylation(GO:0070078) histone H4-R3 demethylation(GO:0070079) |
| 0.1 | 1.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.4 | GO:0002339 | B cell selection(GO:0002339) |
| 0.1 | 0.4 | GO:0019230 | proprioception(GO:0019230) |
| 0.1 | 0.5 | GO:0070889 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.6 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 2.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.7 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.1 | 0.3 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.4 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 1.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.5 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.5 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.1 | 0.6 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 1.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.3 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.6 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.3 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 2.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.3 | GO:1990167 | protein K6-linked deubiquitination(GO:0044313) protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 0.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.6 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.7 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.2 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 0.4 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.1 | 0.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.4 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.4 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.5 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.1 | 0.3 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.3 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.3 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.4 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.3 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.3 | GO:1901536 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.2 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.1 | 0.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.2 | GO:0007174 | epidermal growth factor catabolic process(GO:0007174) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.2 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.3 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.3 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.1 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.0 | 0.5 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.7 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) activation of meiosis(GO:0090427) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.0 | 0.6 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.5 | GO:0072386 | plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.0 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.8 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.5 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.8 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.1 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 0.1 | GO:0003099 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.2 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 3.7 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.3 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.2 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.1 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.0 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.2 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.0 | 1.4 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.2 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.5 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.4 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.3 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.8 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 1.0 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 1.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.0 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.0 | 0.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 1.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.2 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.6 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.4 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.5 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.4 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.1 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.0 | 0.1 | GO:2000843 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.6 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.3 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.0 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.3 | GO:0042346 | positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.0 | 0.1 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.5 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.3 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export(GO:1901660) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.3 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.0 | 1.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.8 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.0 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.1 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.0 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0060455 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:2000504 | regulation of Fas signaling pathway(GO:1902044) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0060846 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) blood vessel endothelial cell fate specification(GO:0097101) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.4 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.4 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.4 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 1.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.0 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.2 | GO:1903350 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.3 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.7 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.6 | 1.8 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.4 | 3.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 1.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.3 | 1.0 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.3 | 0.9 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.2 | 3.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 0.7 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.2 | 2.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 0.5 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.2 | 2.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.5 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.6 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.6 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.4 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.4 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.3 | GO:1990038 | glial cytoplasmic inclusion(GO:0097409) classical Lewy body(GO:0097414) Lewy neurite(GO:0097462) Lewy body corona(GO:1990038) |
| 0.1 | 0.6 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 1.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 6.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.7 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.6 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0001939 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 2.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.6 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 1.7 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 1.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 4.1 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.6 | 4.5 | GO:0023029 | peptide-transporting ATPase activity(GO:0015440) MHC class Ib protein binding(GO:0023029) |
| 0.5 | 1.4 | GO:0016154 | thymidine phosphorylase activity(GO:0009032) pyrimidine-nucleoside phosphorylase activity(GO:0016154) |
| 0.3 | 1.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.3 | 2.5 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.3 | 2.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 0.8 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.2 | 0.6 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.2 | 6.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 2.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.4 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 0.4 | GO:0033746 | histone demethylase activity (H3-R2 specific)(GO:0033746) histone demethylase activity (H4-R3 specific)(GO:0033749) |
| 0.1 | 0.4 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.6 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.8 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.4 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.5 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.9 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.9 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 1.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.3 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 2.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 2.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.9 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.6 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.8 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.8 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 2.0 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.3 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.8 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.1 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.5 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.2 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.3 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 3.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.4 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 1.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 1.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 0.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.7 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.3 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 1.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.2 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.3 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.2 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.0 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.9 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 1.5 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 2.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 2.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 6.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.5 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 1.3 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 4.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 5.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 3.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.3 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 2.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 4.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.2 | REACTOME P75NTR SIGNALS VIA NFKB | Genes involved in p75NTR signals via NF-kB |
| 0.1 | 1.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.6 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 1.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.4 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.1 | 2.0 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 3.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 3.5 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 1.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 2.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 2.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 5.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |