Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
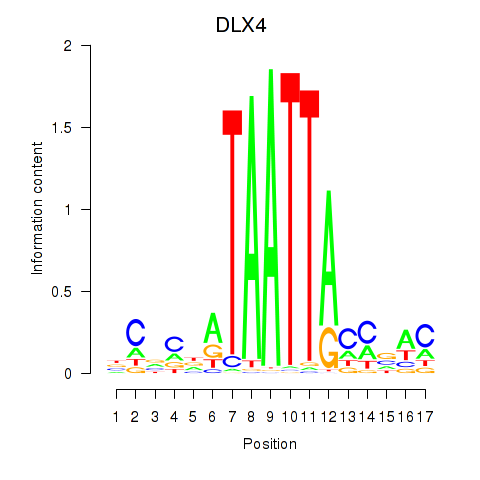
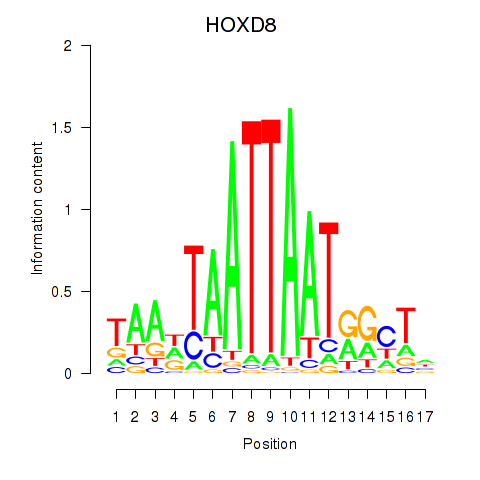
Results for DLX4_HOXD8
Z-value: 1.02
Transcription factors associated with DLX4_HOXD8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX4
|
ENSG00000108813.11 | DLX4 |
|
HOXD8
|
ENSG00000175879.9 | HOXD8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD8 | hg38_v1_chr2_+_176129680_176129764 | -0.32 | 1.2e-01 | Click! |
| DLX4 | hg38_v1_chr17_+_49969178_49969214 | 0.05 | 8.3e-01 | Click! |
Activity profile of DLX4_HOXD8 motif
Sorted Z-values of DLX4_HOXD8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX4_HOXD8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102780620 | 7.82 |
ENST00000279441.9
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 |
| chr2_-_224947030 | 5.36 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr15_+_88635626 | 4.76 |
ENST00000379224.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr10_+_94683771 | 3.72 |
ENST00000339022.6
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr10_+_89392546 | 3.53 |
ENST00000546318.2
ENST00000371804.4 |
IFIT1
|
interferon induced protein with tetratricopeptide repeats 1 |
| chr15_-_79971164 | 3.48 |
ENST00000335661.6
ENST00000267953.4 ENST00000677151.1 |
BCL2A1
|
BCL2 related protein A1 |
| chr12_-_52517929 | 2.96 |
ENST00000548409.5
|
KRT5
|
keratin 5 |
| chr17_-_35880350 | 2.93 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr7_-_116030735 | 2.72 |
ENST00000393485.5
|
TFEC
|
transcription factor EC |
| chr11_+_35180342 | 2.56 |
ENST00000639002.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_-_32964685 | 2.44 |
ENST00000373456.11
ENST00000356990.9 ENST00000235150.5 |
RNF19B
|
ring finger protein 19B |
| chr7_-_116030750 | 2.40 |
ENST00000265440.12
ENST00000320239.11 |
TFEC
|
transcription factor EC |
| chr10_-_88952763 | 2.39 |
ENST00000224784.10
|
ACTA2
|
actin alpha 2, smooth muscle |
| chr11_+_35180279 | 2.35 |
ENST00000531873.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr18_+_58341038 | 2.13 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr7_-_93148345 | 2.10 |
ENST00000437805.5
ENST00000446959.5 ENST00000439952.5 ENST00000414791.5 ENST00000446033.1 ENST00000411955.5 ENST00000318238.9 |
SAMD9L
|
sterile alpha motif domain containing 9 like |
| chr22_+_18150162 | 1.98 |
ENST00000215794.8
|
USP18
|
ubiquitin specific peptidase 18 |
| chr2_+_102418642 | 1.90 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr1_-_120054225 | 1.89 |
ENST00000602566.6
|
NOTCH2
|
notch receptor 2 |
| chr14_-_91253925 | 1.79 |
ENST00000531499.2
|
GPR68
|
G protein-coupled receptor 68 |
| chr21_-_14546297 | 1.75 |
ENST00000400566.6
ENST00000400564.5 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr8_+_12104389 | 1.71 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D |
| chr5_-_36301883 | 1.70 |
ENST00000502994.5
ENST00000515759.5 ENST00000296604.8 |
RANBP3L
|
RAN binding protein 3 like |
| chr11_+_5689780 | 1.54 |
ENST00000379965.8
ENST00000454828.5 |
TRIM22
|
tripartite motif containing 22 |
| chr5_+_157269317 | 1.50 |
ENST00000618329.4
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr3_+_122680802 | 1.47 |
ENST00000474629.7
|
PARP14
|
poly(ADP-ribose) polymerase family member 14 |
| chr5_-_160852200 | 1.47 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr12_+_112907006 | 1.42 |
ENST00000680455.1
ENST00000551241.6 ENST00000550689.2 ENST00000679841.1 ENST00000679494.1 ENST00000553185.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1 |
| chr3_+_155083889 | 1.41 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase |
| chr1_-_89126066 | 1.39 |
ENST00000370466.4
|
GBP2
|
guanylate binding protein 2 |
| chr4_-_69214743 | 1.36 |
ENST00000446444.2
|
UGT2B11
|
UDP glucuronosyltransferase family 2 member B11 |
| chr5_-_59216826 | 1.34 |
ENST00000638939.1
|
PDE4D
|
phosphodiesterase 4D |
| chr16_-_28610032 | 1.34 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr6_-_52840843 | 1.34 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr8_-_89984609 | 1.31 |
ENST00000519426.5
ENST00000265433.8 |
NBN
|
nibrin |
| chr6_+_29301701 | 1.31 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr12_+_112906949 | 1.31 |
ENST00000679971.1
ENST00000675868.2 ENST00000550883.2 ENST00000553152.2 ENST00000202917.10 ENST00000679467.1 ENST00000680659.1 ENST00000540589.3 ENST00000552526.2 ENST00000681228.1 ENST00000680934.1 ENST00000681700.1 ENST00000679987.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1 |
| chr21_-_14546351 | 1.26 |
ENST00000619120.4
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr4_-_68670648 | 1.26 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr1_-_197146620 | 1.23 |
ENST00000367409.9
ENST00000680265.1 |
ASPM
|
assembly factor for spindle microtubules |
| chr12_-_91179355 | 1.22 |
ENST00000550563.5
ENST00000546370.5 |
DCN
|
decorin |
| chr12_+_9827472 | 1.22 |
ENST00000617793.4
ENST00000617889.5 ENST00000354855.7 ENST00000279545.7 |
KLRF1
|
killer cell lectin like receptor F1 |
| chr8_+_103298433 | 1.21 |
ENST00000522566.5
|
FZD6
|
frizzled class receptor 6 |
| chr16_-_28623560 | 1.20 |
ENST00000350842.8
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr11_-_117877463 | 1.20 |
ENST00000527717.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr12_+_26195313 | 1.18 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr6_-_87095059 | 1.16 |
ENST00000369582.6
ENST00000610310.3 ENST00000630630.2 ENST00000627148.3 ENST00000625577.1 |
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr6_+_32844789 | 1.15 |
ENST00000414474.5
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr2_+_151357583 | 1.15 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6 |
| chr18_-_36122110 | 1.12 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 member 6 |
| chr1_-_72100930 | 1.10 |
ENST00000306821.3
|
NEGR1
|
neuronal growth regulator 1 |
| chr1_-_150765785 | 1.08 |
ENST00000680311.1
ENST00000681728.1 ENST00000680288.1 |
CTSS
|
cathepsin S |
| chr1_+_78649818 | 1.08 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr3_+_155083523 | 1.08 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase |
| chr8_-_89984231 | 1.07 |
ENST00000517337.1
ENST00000409330.5 |
NBN
|
nibrin |
| chr11_+_20022550 | 1.06 |
ENST00000533917.5
|
NAV2
|
neuron navigator 2 |
| chr3_-_191282383 | 1.05 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr1_-_205121986 | 1.04 |
ENST00000367164.1
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr2_+_167135901 | 1.03 |
ENST00000628543.2
|
XIRP2
|
xin actin binding repeat containing 2 |
| chr7_-_115968302 | 1.02 |
ENST00000457268.5
|
TFEC
|
transcription factor EC |
| chr3_-_79767987 | 1.02 |
ENST00000464233.6
|
ROBO1
|
roundabout guidance receptor 1 |
| chr8_-_85341705 | 1.00 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr1_-_247458105 | 1.00 |
ENST00000641149.1
ENST00000641527.1 |
OR2B11
|
olfactory receptor family 2 subfamily B member 11 |
| chr1_+_86547070 | 1.00 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr4_+_73740541 | 0.99 |
ENST00000401931.1
ENST00000307407.8 |
CXCL8
|
C-X-C motif chemokine ligand 8 |
| chr1_-_197146688 | 0.97 |
ENST00000294732.11
|
ASPM
|
assembly factor for spindle microtubules |
| chr11_+_18412292 | 0.97 |
ENST00000541669.6
ENST00000280704.8 |
LDHC
|
lactate dehydrogenase C |
| chr1_-_205121964 | 0.97 |
ENST00000264515.11
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr21_+_42199686 | 0.96 |
ENST00000398457.6
|
ABCG1
|
ATP binding cassette subfamily G member 1 |
| chr20_-_1657714 | 0.96 |
ENST00000216927.4
ENST00000344103.8 |
SIRPG
|
signal regulatory protein gamma |
| chr2_-_201698040 | 0.96 |
ENST00000396886.7
ENST00000409143.5 |
MPP4
|
membrane palmitoylated protein 4 |
| chr4_-_69653223 | 0.96 |
ENST00000286604.8
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
|
UDP glucuronosyltransferase family 2 member A1 complex locus |
| chr22_+_20774092 | 0.96 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chr12_-_122500520 | 0.95 |
ENST00000540586.1
ENST00000543897.5 |
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr1_-_247758680 | 0.95 |
ENST00000408896.4
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1 |
| chr10_-_77637902 | 0.94 |
ENST00000286627.10
ENST00000639486.1 ENST00000640523.1 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr12_+_9669735 | 0.94 |
ENST00000545918.5
ENST00000543300.5 ENST00000261339.10 ENST00000466035.6 |
CLEC2D
|
C-type lectin domain family 2 member D |
| chr11_-_4393650 | 0.94 |
ENST00000254436.8
|
TRIM21
|
tripartite motif containing 21 |
| chr11_-_117876719 | 0.93 |
ENST00000529335.6
ENST00000260282.8 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr2_-_89213917 | 0.93 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr7_+_138460238 | 0.93 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr21_-_14658812 | 0.93 |
ENST00000647101.1
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr12_-_9999176 | 0.92 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr15_+_76336755 | 0.91 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr11_+_35176611 | 0.91 |
ENST00000279452.10
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_+_55811367 | 0.90 |
ENST00000625203.2
|
OR5L1
|
olfactory receptor family 5 subfamily L member 1 |
| chr10_+_80131660 | 0.89 |
ENST00000372270.6
|
PLAC9
|
placenta associated 9 |
| chr1_-_89065200 | 0.89 |
ENST00000370473.5
|
GBP1
|
guanylate binding protein 1 |
| chr5_-_20575850 | 0.89 |
ENST00000507958.5
|
CDH18
|
cadherin 18 |
| chr8_+_27774566 | 0.89 |
ENST00000519637.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr14_-_67412112 | 0.88 |
ENST00000216446.9
|
PLEK2
|
pleckstrin 2 |
| chr1_+_158461574 | 0.87 |
ENST00000641432.1
ENST00000641460.1 ENST00000641535.1 ENST00000641971.1 |
OR10K1
|
olfactory receptor family 10 subfamily K member 1 |
| chr9_-_21482313 | 0.86 |
ENST00000448696.4
|
IFNE
|
interferon epsilon |
| chr4_-_48080172 | 0.86 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr1_-_88891496 | 0.86 |
ENST00000448623.5
ENST00000370500.10 ENST00000418217.1 |
GTF2B
|
general transcription factor IIB |
| chr8_+_103372388 | 0.86 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr11_-_117876892 | 0.85 |
ENST00000539526.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_+_56027654 | 0.84 |
ENST00000641320.1
|
OR5AS1
|
olfactory receptor family 5 subfamily AS member 1 |
| chr7_-_97872420 | 0.84 |
ENST00000444334.5
ENST00000422745.5 ENST00000451771.5 ENST00000175506.8 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr1_+_119711884 | 0.84 |
ENST00000641947.1
ENST00000641074.1 ENST00000641115.1 ENST00000369409.9 ENST00000641023.2 ENST00000641272.1 |
PHGDH
|
phosphoglycerate dehydrogenase |
| chr17_-_31901658 | 0.84 |
ENST00000261708.9
|
UTP6
|
UTP6 small subunit processome component |
| chr4_-_76007501 | 0.82 |
ENST00000264888.6
|
CXCL9
|
C-X-C motif chemokine ligand 9 |
| chr1_-_247760556 | 0.82 |
ENST00000641256.1
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1 |
| chr11_+_35176575 | 0.82 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_74235377 | 0.81 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr17_+_42798779 | 0.81 |
ENST00000585355.5
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr10_-_122845850 | 0.80 |
ENST00000392790.6
|
CUZD1
|
CUB and zona pellucida like domains 1 |
| chr13_-_110242694 | 0.80 |
ENST00000648989.1
ENST00000647797.1 ENST00000648966.1 ENST00000649484.1 ENST00000648695.1 ENST00000650115.1 ENST00000650566.1 |
COL4A1
|
collagen type IV alpha 1 chain |
| chr13_-_101416441 | 0.79 |
ENST00000675332.1
ENST00000676315.1 ENST00000251127.11 |
NALCN
|
sodium leak channel, non-selective |
| chr3_+_155080307 | 0.79 |
ENST00000360490.7
|
MME
|
membrane metalloendopeptidase |
| chr4_+_122732652 | 0.79 |
ENST00000542236.5
ENST00000314218.8 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr10_+_55599041 | 0.79 |
ENST00000512524.4
|
MTRNR2L5
|
MT-RNR2 like 5 |
| chr11_+_35176696 | 0.78 |
ENST00000528455.5
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_+_124185216 | 0.78 |
ENST00000318666.6
|
OR10D3
|
olfactory receptor family 10 subfamily D member 3 |
| chr5_+_122129533 | 0.78 |
ENST00000296600.5
ENST00000504912.1 ENST00000505843.1 |
ZNF474
|
zinc finger protein 474 |
| chr19_-_3557563 | 0.78 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr4_-_159035226 | 0.78 |
ENST00000434826.3
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr5_-_170389349 | 0.77 |
ENST00000274629.9
|
KCNMB1
|
potassium calcium-activated channel subfamily M regulatory beta subunit 1 |
| chr20_-_36951837 | 0.77 |
ENST00000262878.5
|
SAMHD1
|
SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 |
| chr14_-_52069039 | 0.76 |
ENST00000216286.10
|
NID2
|
nidogen 2 |
| chr17_-_66229380 | 0.76 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr19_+_53962925 | 0.76 |
ENST00000270458.4
|
CACNG8
|
calcium voltage-gated channel auxiliary subunit gamma 8 |
| chr2_-_201698692 | 0.76 |
ENST00000315506.11
ENST00000359962.9 ENST00000620095.4 |
MPP4
|
membrane palmitoylated protein 4 |
| chr22_-_36160773 | 0.76 |
ENST00000531095.1
ENST00000349314.6 ENST00000397293.6 |
APOL3
|
apolipoprotein L3 |
| chr11_-_114595777 | 0.76 |
ENST00000375478.4
|
NXPE4
|
neurexophilin and PC-esterase domain family member 4 |
| chr12_-_119803383 | 0.75 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr14_-_52069228 | 0.75 |
ENST00000617139.4
|
NID2
|
nidogen 2 |
| chr21_+_25639251 | 0.75 |
ENST00000480456.6
|
JAM2
|
junctional adhesion molecule 2 |
| chr6_-_37258110 | 0.74 |
ENST00000357219.4
ENST00000652386.1 ENST00000652639.1 |
TMEM217
|
transmembrane protein 217 |
| chr16_-_75556214 | 0.74 |
ENST00000568377.5
ENST00000565067.5 ENST00000258173.11 |
TMEM231
|
transmembrane protein 231 |
| chr8_-_124565699 | 0.74 |
ENST00000519168.5
|
MTSS1
|
MTSS I-BAR domain containing 1 |
| chr4_+_109815503 | 0.74 |
ENST00000394631.7
|
GAR1
|
GAR1 ribonucleoprotein |
| chr12_+_14419136 | 0.73 |
ENST00000545769.1
ENST00000428217.6 ENST00000396279.2 ENST00000542514.5 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr21_-_36480060 | 0.73 |
ENST00000399137.5
|
CLDN14
|
claudin 14 |
| chr21_+_25639272 | 0.72 |
ENST00000400532.5
ENST00000312957.9 |
JAM2
|
junctional adhesion molecule 2 |
| chr3_-_112829367 | 0.72 |
ENST00000448932.4
ENST00000617549.3 |
CD200R1L
|
CD200 receptor 1 like |
| chr9_+_5510492 | 0.72 |
ENST00000397747.5
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr2_+_89884740 | 0.72 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr8_+_12108172 | 0.71 |
ENST00000400078.3
|
ZNF705D
|
zinc finger protein 705D |
| chr11_-_102625332 | 0.70 |
ENST00000260228.3
|
MMP20
|
matrix metallopeptidase 20 |
| chr11_-_55936400 | 0.70 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr5_-_97183203 | 0.70 |
ENST00000508447.1
ENST00000283109.8 |
RIOK2
|
RIO kinase 2 |
| chr10_+_58269132 | 0.70 |
ENST00000333926.6
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr17_+_42980547 | 0.69 |
ENST00000361677.5
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr13_+_50909905 | 0.68 |
ENST00000644034.1
ENST00000645955.1 |
RNASEH2B
|
ribonuclease H2 subunit B |
| chr5_+_129748091 | 0.68 |
ENST00000564719.2
|
MINAR2
|
membrane integral NOTCH2 associated receptor 2 |
| chr3_-_132684685 | 0.68 |
ENST00000512094.5
ENST00000632629.1 |
NPHP3
NPHP3-ACAD11
|
nephrocystin 3 NPHP3-ACAD11 readthrough (NMD candidate) |
| chr6_-_11779606 | 0.68 |
ENST00000506810.1
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr3_-_12545499 | 0.67 |
ENST00000564146.4
|
MKRN2OS
|
MKRN2 opposite strand |
| chr18_+_24113341 | 0.67 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr8_-_13276491 | 0.67 |
ENST00000512044.6
|
DLC1
|
DLC1 Rho GTPase activating protein |
| chr4_-_39977836 | 0.66 |
ENST00000303538.13
ENST00000503396.5 |
PDS5A
|
PDS5 cohesin associated factor A |
| chr5_-_93741587 | 0.66 |
ENST00000606183.4
|
POU5F2
|
POU domain class 5, transcription factor 2 |
| chr6_-_57221402 | 0.65 |
ENST00000317483.4
|
RAB23
|
RAB23, member RAS oncogene family |
| chr1_-_158686700 | 0.65 |
ENST00000643759.2
|
SPTA1
|
spectrin alpha, erythrocytic 1 |
| chr11_+_35176639 | 0.64 |
ENST00000527889.6
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_-_35360050 | 0.64 |
ENST00000644868.1
ENST00000643454.1 ENST00000646080.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr17_-_69150062 | 0.64 |
ENST00000522787.5
ENST00000521538.5 |
ABCA10
|
ATP binding cassette subfamily A member 10 |
| chr12_-_89526011 | 0.64 |
ENST00000313546.8
|
POC1B
|
POC1 centriolar protein B |
| chr19_+_984314 | 0.64 |
ENST00000587001.6
ENST00000585809.6 ENST00000251289.9 ENST00000607440.5 |
WDR18
|
WD repeat domain 18 |
| chr11_-_72794032 | 0.64 |
ENST00000334805.11
|
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr7_+_142791635 | 0.63 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1 |
| chr8_-_61646807 | 0.63 |
ENST00000522919.5
|
ASPH
|
aspartate beta-hydroxylase |
| chr4_+_74308463 | 0.63 |
ENST00000413830.6
|
EPGN
|
epithelial mitogen |
| chr9_-_83267230 | 0.63 |
ENST00000328788.5
|
FRMD3
|
FERM domain containing 3 |
| chr1_+_206635521 | 0.63 |
ENST00000367109.8
|
DYRK3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr12_-_10909562 | 0.63 |
ENST00000390677.2
|
TAS2R13
|
taste 2 receptor member 13 |
| chrX_+_30243715 | 0.62 |
ENST00000378981.8
ENST00000397550.6 |
MAGEB1
|
MAGE family member B1 |
| chr15_+_67125707 | 0.62 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr11_+_124241095 | 0.62 |
ENST00000641972.1
|
OR8G1
|
olfactory receptor family 8 subfamily G member 1 |
| chr16_-_28609992 | 0.62 |
ENST00000314752.11
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chr6_+_116399395 | 0.62 |
ENST00000644499.1
|
ENSG00000285446.1
|
novel protein |
| chr21_-_36480117 | 0.61 |
ENST00000399135.6
|
CLDN14
|
claudin 14 |
| chr3_+_173398438 | 0.61 |
ENST00000457714.5
|
NLGN1
|
neuroligin 1 |
| chr10_+_7703340 | 0.61 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr15_+_71096941 | 0.61 |
ENST00000355327.7
|
THSD4
|
thrombospondin type 1 domain containing 4 |
| chr21_-_30813270 | 0.61 |
ENST00000329621.6
|
KRTAP8-1
|
keratin associated protein 8-1 |
| chr12_+_21437642 | 0.60 |
ENST00000240651.14
|
PYROXD1
|
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr11_+_35186820 | 0.60 |
ENST00000531110.6
ENST00000525685.6 |
CD44
|
CD44 molecule (Indian blood group) |
| chrX_-_15314543 | 0.59 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr20_+_33217325 | 0.59 |
ENST00000375452.3
ENST00000375454.8 |
BPIFA3
|
BPI fold containing family A member 3 |
| chr10_+_88664439 | 0.59 |
ENST00000394375.7
ENST00000608620.5 ENST00000238983.9 ENST00000355843.2 |
LIPF
|
lipase F, gastric type |
| chr2_-_89117844 | 0.59 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr3_+_149474688 | 0.59 |
ENST00000305354.5
ENST00000465758.1 |
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr1_+_172420681 | 0.58 |
ENST00000367727.9
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr7_-_44082464 | 0.58 |
ENST00000335195.10
ENST00000395831.7 ENST00000414235.5 ENST00000242248.10 ENST00000452049.1 |
POLM
|
DNA polymerase mu |
| chr6_-_28443463 | 0.58 |
ENST00000289788.4
|
ZSCAN23
|
zinc finger and SCAN domain containing 23 |
| chr6_-_118710065 | 0.57 |
ENST00000392500.7
ENST00000368488.9 ENST00000434604.5 |
CEP85L
|
centrosomal protein 85 like |
| chr10_-_77637789 | 0.57 |
ENST00000481070.1
ENST00000640969.1 ENST00000286628.14 ENST00000638991.1 ENST00000639913.1 ENST00000480683.2 |
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr7_-_149461056 | 0.57 |
ENST00000247930.5
|
ZNF777
|
zinc finger protein 777 |
| chr1_-_36440873 | 0.57 |
ENST00000433045.6
|
OSCP1
|
organic solute carrier partner 1 |
| chr4_+_174918355 | 0.57 |
ENST00000505141.5
ENST00000359240.7 ENST00000615367.4 ENST00000445694.5 ENST00000618444.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr11_-_57567617 | 0.56 |
ENST00000287156.9
|
UBE2L6
|
ubiquitin conjugating enzyme E2 L6 |
| chr10_-_113854368 | 0.56 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr3_-_190122317 | 0.56 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chrX_-_77969638 | 0.56 |
ENST00000458128.3
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr4_-_79326008 | 0.56 |
ENST00000286794.5
|
NAA11
|
N-alpha-acetyltransferase 11, NatA catalytic subunit |
| chr4_+_87832917 | 0.55 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr3_-_112846856 | 0.55 |
ENST00000488794.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr7_+_151341764 | 0.54 |
ENST00000413040.7
ENST00000470229.6 ENST00000568733.6 |
NUB1
|
negative regulator of ubiquitin like proteins 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.2 | 3.5 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.7 | 2.9 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.7 | 8.7 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.6 | 2.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.5 | 2.4 | GO:0090131 | glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.5 | 0.9 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.4 | 7.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.4 | 2.8 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.4 | 1.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.3 | 1.4 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.3 | 3.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 1.0 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.3 | 0.9 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.3 | 1.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.3 | 0.8 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.3 | 0.6 | GO:0031393 | negative regulation of prostaglandin biosynthetic process(GO:0031393) |
| 0.3 | 1.0 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of negative chemotaxis(GO:0050925) |
| 0.2 | 2.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.2 | 0.9 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.2 | 0.8 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.2 | 0.8 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 2.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 1.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 2.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 1.7 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 0.6 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 | 4.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 1.1 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.2 | 1.0 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.2 | 0.9 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.2 | 0.7 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 0.5 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.2 | 2.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.2 | 0.7 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 0.8 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 1.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 1.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 0.6 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.1 | 0.4 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.1 | 0.9 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.7 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.4 | GO:0048058 | compound eye corneal lens development(GO:0048058) |
| 0.1 | 0.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.9 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.1 | 0.6 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.6 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.8 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.4 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.7 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 1.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 4.7 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.4 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.4 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.1 | 0.9 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.6 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.1 | 0.5 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.4 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.1 | 1.4 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 0.5 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 0.5 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.5 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.8 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.5 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.1 | 0.7 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.4 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.1 | 0.5 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 3.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.6 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.5 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.6 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 0.5 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 9.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.8 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.4 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.1 | 1.0 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 0.8 | GO:0046061 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.1 | 0.4 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 1.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.6 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 3.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.4 | GO:0090095 | regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.3 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.1 | 1.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.4 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 1.0 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 1.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.6 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.3 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.1 | 0.7 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.9 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.2 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.3 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.5 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.5 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.3 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.5 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.5 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 0.2 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.8 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.2 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 3.6 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.8 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.1 | 0.2 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.1 | 0.2 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 0.8 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.3 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.1 | 0.3 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.4 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.2 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.1 | 0.1 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.9 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 1.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.2 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 0.2 | GO:0035691 | macrophage migration inhibitory factor signaling pathway(GO:0035691) |
| 0.1 | 0.3 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.1 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) |
| 0.1 | 0.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.1 | 0.4 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.1 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 | 0.2 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.1 | 0.2 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 0.4 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.2 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.2 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 10.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.3 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.7 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 0.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.4 | GO:0042262 | DNA protection(GO:0042262) |
| 0.1 | 1.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.3 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 0.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.1 | 0.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 1.3 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.6 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.4 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.3 | GO:0089712 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.0 | 0.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 1.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.9 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.4 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.6 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.3 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.2 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.4 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.2 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.3 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.2 | GO:0070889 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.2 | GO:0019376 | galactolipid catabolic process(GO:0019376) |
| 0.0 | 0.1 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.6 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0060739 | mesenchymal-epithelial cell signaling involved in prostate gland development(GO:0060739) |
| 0.0 | 0.6 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.4 | GO:1903943 | negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 3.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.3 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.4 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.4 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 2.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0046469 | platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.3 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.3 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.2 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 | 0.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.3 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.5 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 2.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.5 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.1 | GO:0009189 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.6 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) |
| 0.0 | 1.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.1 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 2.0 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.1 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.7 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.4 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 5.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.2 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) Golgi disassembly(GO:0090166) |
| 0.0 | 0.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 1.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:1903546 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.2 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.5 | GO:0036314 | response to sterol(GO:0036314) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.3 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.0 | 0.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.0 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.6 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.4 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 2.5 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 1.0 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.9 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.0 | 0.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.6 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0007174 | epidermal growth factor catabolic process(GO:0007174) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 2.1 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.1 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.2 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 1.7 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:1902688 | regulation of NAD metabolic process(GO:1902688) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0097502 | mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.3 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.1 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.4 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.2 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.2 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.0 | 0.5 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.0 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.2 | GO:0043622 | cortical microtubule organization(GO:0043622) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.5 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.0 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 1.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.3 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.3 | GO:0009309 | amine biosynthetic process(GO:0009309) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.0 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.0 | GO:0038001 | paracrine signaling(GO:0038001) |
| 0.0 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.2 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 0.0 | 0.8 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.2 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.0 | GO:0015744 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 8.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 2.0 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 2.4 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 0.7 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 2.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 0.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 2.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 1.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 0.5 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.2 | 1.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 2.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.0 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.4 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 0.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.7 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.8 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.3 | GO:0071752 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) |
| 0.1 | 0.4 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.3 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.1 | 6.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.2 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.3 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.2 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.5 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 4.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.0 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 1.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 1.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 3.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.0 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 1.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.4 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 2.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 3.0 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 1.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 6.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 5.4 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 6.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.0 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network(GO:0031094) platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0032419 | extrinsic component of lysosome membrane(GO:0032419) |
| 0.0 | 8.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 1.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.3 | GO:0043073 | male germ cell nucleus(GO:0001673) germ cell nucleus(GO:0043073) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 1.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 1.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 2.1 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.1 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 1.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.7 | 3.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.7 | 4.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.3 | 0.8 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.2 | 9.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 1.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.2 | 0.7 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.2 | 2.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 4.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 1.9 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.2 | 6.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 2.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 0.8 | GO:0032560 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) guanyl deoxyribonucleotide binding(GO:0032560) dGTP binding(GO:0032567) |
| 0.2 | 0.6 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.2 | 3.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 0.5 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.2 | 0.8 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 0.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 1.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.2 | 0.5 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.2 | 0.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.4 | GO:0001034 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.1 | 0.8 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.4 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.6 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.6 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 1.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.6 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.5 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 1.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.5 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.4 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 1.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.6 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.5 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.6 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.7 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 1.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.9 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.6 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.6 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 2.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.5 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.6 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 0.3 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 15.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.5 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 2.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.3 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.1 | 0.8 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 1.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.2 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.3 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.4 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.1 | 0.4 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 4.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 6.2 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 2.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 1.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.4 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.1 | 0.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 2.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 1.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.2 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 0.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.3 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.3 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 1.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 2.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.9 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 1.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.4 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 4.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.0 | GO:0052854 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.6 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0097027 | anaphase-promoting complex binding(GO:0010997) ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0015563 | thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.5 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 5.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 2.0 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 4.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 8.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 3.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 3.7 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 14.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 4.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 7.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 10.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 10.5 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.3 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.1 | 0.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 3.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 2.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.3 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 1.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 2.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.6 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |