Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
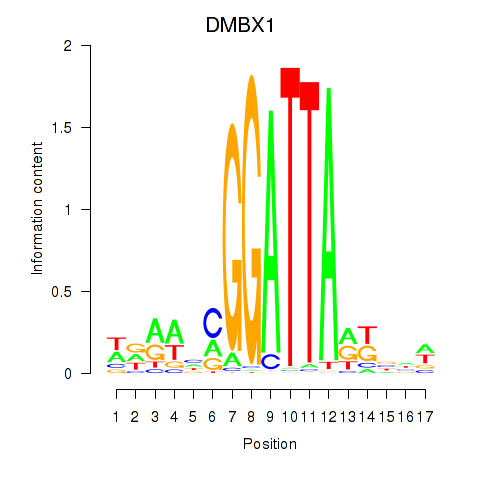
Results for DMBX1
Z-value: 0.38
Transcription factors associated with DMBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMBX1
|
ENSG00000197587.11 | DMBX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DMBX1 | hg38_v1_chr1_+_46489800_46489842 | -0.36 | 7.6e-02 | Click! |
Activity profile of DMBX1 motif
Sorted Z-values of DMBX1 motif
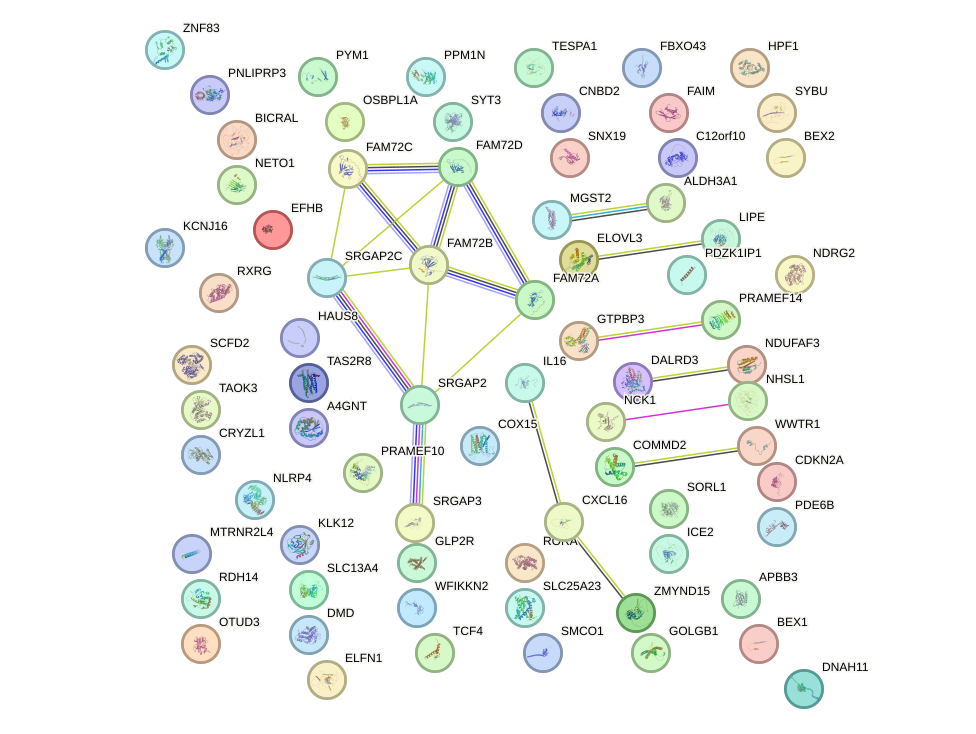
Network of associatons between targets according to the STRING database.
First level regulatory network of DMBX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_44680875 | 0.93 |
ENST00000649279.2
ENST00000677699.1 |
SETBP1
|
SET binding protein 1 |
| chr4_+_139665768 | 0.70 |
ENST00000616265.4
ENST00000265498.6 ENST00000506797.5 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr3_-_138132381 | 0.65 |
ENST00000236709.4
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr2_-_70553440 | 0.48 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha |
| chr12_-_56333893 | 0.40 |
ENST00000547572.1
ENST00000257931.9 ENST00000610546.4 ENST00000440411.7 |
PAN2
|
poly(A) specific ribonuclease subunit PAN2 |
| chr19_+_45498439 | 0.39 |
ENST00000451287.7
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative) |
| chrX_-_103311004 | 0.39 |
ENST00000372674.5
|
BEX2
|
brain expressed X-linked 2 |
| chr10_+_102226293 | 0.38 |
ENST00000370005.4
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr12_-_56333693 | 0.35 |
ENST00000425394.7
ENST00000548043.5 |
PAN2
|
poly(A) specific ribonuclease subunit PAN2 |
| chr6_+_130018565 | 0.34 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr16_-_3372666 | 0.34 |
ENST00000399974.5
|
MTRNR2L4
|
MT-RNR2 like 4 |
| chr11_+_121576760 | 0.34 |
ENST00000532694.5
ENST00000534286.5 |
SORL1
|
sortilin related receptor 1 |
| chr3_-_49021045 | 0.33 |
ENST00000440857.5
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr1_-_165445088 | 0.30 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma |
| chr1_-_165445220 | 0.30 |
ENST00000619224.1
|
RXRG
|
retinoid X receptor gamma |
| chr4_-_53365976 | 0.29 |
ENST00000401642.8
ENST00000388940.8 ENST00000503450.1 |
SCFD2
|
sec1 family domain containing 2 |
| chr17_+_4740042 | 0.28 |
ENST00000592813.5
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr17_+_4740005 | 0.28 |
ENST00000269289.10
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr19_-_6459735 | 0.28 |
ENST00000334510.9
ENST00000301454.9 |
SLC25A23
|
solute carrier family 25 member 23 |
| chr19_+_40778216 | 0.27 |
ENST00000594800.5
ENST00000357052.8 ENST00000602173.5 |
RAB4B
|
RAB4B, member RAS oncogene family |
| chr3_-_149576203 | 0.27 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr3_+_136930469 | 0.25 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr12_+_53300027 | 0.24 |
ENST00000549488.5
|
MYG1
|
MYG1 exonuclease |
| chr3_-_121749704 | 0.24 |
ENST00000393667.7
ENST00000340645.9 ENST00000614479.4 |
GOLGB1
|
golgin B1 |
| chr18_-_55587335 | 0.23 |
ENST00000638154.3
|
TCF4
|
transcription factor 4 |
| chr1_+_19882374 | 0.22 |
ENST00000375120.4
|
OTUD3
|
OTU deubiquitinase 3 |
| chr4_-_169757873 | 0.22 |
ENST00000393381.3
|
HPF1
|
histone PARylation factor 1 |
| chr12_-_10807286 | 0.22 |
ENST00000240615.3
|
TAS2R8
|
taste 2 receptor member 8 |
| chr12_-_118190510 | 0.19 |
ENST00000540561.5
ENST00000537952.1 ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr10_-_99732070 | 0.18 |
ENST00000370483.9
ENST00000016171.6 |
COX15
|
cytochrome c oxidase assembly homolog COX15 |
| chr7_+_1688119 | 0.18 |
ENST00000424383.4
|
ELFN1
|
extracellular leucine rich repeat and fibronectin type III domain containing 1 |
| chr15_-_60479080 | 0.17 |
ENST00000560072.5
ENST00000560406.5 ENST00000560520.1 ENST00000261520.9 |
ICE2
|
interactor of little elongation complex ELL subunit 2 |
| chr12_-_54981838 | 0.16 |
ENST00000316577.12
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr6_+_42746958 | 0.16 |
ENST00000614467.4
|
BICRAL
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr17_+_50834581 | 0.15 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr3_+_138621225 | 0.14 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr4_+_625555 | 0.14 |
ENST00000496514.6
ENST00000255622.10 |
PDE6B
|
phosphodiesterase 6B |
| chr17_-_4739866 | 0.14 |
ENST00000574412.6
ENST00000293778.12 |
CXCL16
|
C-X-C motif chemokine ligand 16 |
| chrX_-_32155462 | 0.14 |
ENST00000359836.5
ENST00000378707.7 ENST00000541735.5 ENST00000684130.1 ENST00000682238.1 ENST00000620040.5 ENST00000474231.5 |
DMD
|
dystrophin |
| chr2_-_207624983 | 0.13 |
ENST00000448007.6
ENST00000432416.5 ENST00000411432.5 ENST00000458426.5 ENST00000406927.6 ENST00000425132.5 |
METTL21A
|
methyltransferase like 21A |
| chr19_-_50637939 | 0.12 |
ENST00000338916.8
|
SYT3
|
synaptotagmin 3 |
| chr19_-_17075418 | 0.12 |
ENST00000253669.10
|
HAUS8
|
HAUS augmin like complex subunit 8 |
| chr3_-_19934189 | 0.11 |
ENST00000295824.14
|
EFHB
|
EF-hand domain family member B |
| chr19_-_17075038 | 0.11 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin like complex subunit 8 |
| chr19_-_42412347 | 0.11 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase E, hormone sensitive type |
| chrX_-_15854791 | 0.11 |
ENST00000545766.7
ENST00000380291.5 ENST00000672987.1 ENST00000329235.6 |
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chr1_+_121184964 | 0.10 |
ENST00000367123.8
|
SRGAP2C
|
SLIT-ROBO Rho GTPase activating protein 2C |
| chr21_-_33643926 | 0.10 |
ENST00000438788.1
|
CRYZL1
|
crystallin zeta like 1 |
| chr1_-_145095528 | 0.10 |
ENST00000612199.4
ENST00000641863.1 |
SRGAP2B
|
SLIT-ROBO Rho GTPase activating protein 2B |
| chr17_-_19748341 | 0.09 |
ENST00000395555.7
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr17_-_19748285 | 0.09 |
ENST00000570414.1
ENST00000225740.11 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr3_-_149752443 | 0.09 |
ENST00000473414.6
|
COMMD2
|
COMM domain containing 2 |
| chr17_-_19748355 | 0.09 |
ENST00000494157.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr3_-_196515315 | 0.09 |
ENST00000397537.3
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr1_-_121184292 | 0.09 |
ENST00000452190.2
ENST00000619376.4 ENST00000369390.7 |
FAM72B
|
family with sequence similarity 72 member B |
| chr1_-_143971965 | 0.09 |
ENST00000369175.4
ENST00000584486.6 |
FAM72C
|
family with sequence similarity 72 member C |
| chr1_-_206202827 | 0.08 |
ENST00000431655.2
ENST00000367128.8 |
FAM72A
|
family with sequence similarity 72 member A |
| chr13_-_61427849 | 0.08 |
ENST00000409186.1
ENST00000472649.2 |
ENSG00000197991.11
LINC02339
|
novel protein long intergenic non-protein coding RNA 2339 |
| chr1_+_206203541 | 0.08 |
ENST00000573034.8
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr15_-_61229297 | 0.08 |
ENST00000335670.11
|
RORA
|
RAR related orphan receptor A |
| chr3_+_138621207 | 0.08 |
ENST00000464668.5
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr8_-_100145802 | 0.08 |
ENST00000428847.3
|
FBXO43
|
F-box protein 43 |
| chr1_+_145095967 | 0.07 |
ENST00000400889.3
|
FAM72D
|
family with sequence similarity 72 member D |
| chr7_-_135728177 | 0.07 |
ENST00000682651.1
ENST00000354042.8 |
SLC13A4
|
solute carrier family 13 member 4 |
| chr18_-_24272179 | 0.07 |
ENST00000399443.7
|
OSBPL1A
|
oxysterol binding protein like 1A |
| chr5_+_176313080 | 0.06 |
ENST00000332772.4
|
SIMC1
|
SUMO interacting motifs containing 1 |
| chr11_-_130916437 | 0.06 |
ENST00000533214.1
ENST00000528555.5 ENST00000530356.5 ENST00000265909.9 |
SNX19
|
sorting nexin 19 |
| chr14_-_106875069 | 0.06 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr1_-_13347134 | 0.05 |
ENST00000334600.7
|
PRAMEF14
|
PRAME family member 14 |
| chr10_+_116427839 | 0.05 |
ENST00000369230.4
|
PNLIPRP3
|
pancreatic lipase related protein 3 |
| chr14_-_21022817 | 0.05 |
ENST00000554104.5
|
NDRG2
|
NDRG family member 2 |
| chr5_-_140564245 | 0.05 |
ENST00000412920.7
ENST00000511201.2 ENST00000354402.9 ENST00000356738.6 |
APBB3
|
amyloid beta precursor protein binding family B member 3 |
| chr7_+_29122274 | 0.04 |
ENST00000582692.2
ENST00000644824.1 |
ENSG00000285412.1
ENSG00000285162.1
|
novel transcript, antisense to CPVL chimerin 2 |
| chr19_+_17337539 | 0.04 |
ENST00000324894.13
ENST00000358792.11 ENST00000600625.5 |
GTPBP3
|
GTP binding protein 3, mitochondrial |
| chr15_+_81182579 | 0.03 |
ENST00000302987.9
|
IL16
|
interleukin 16 |
| chr12_-_55927865 | 0.03 |
ENST00000454792.2
ENST00000408946.7 |
PYM1
|
PYM homolog 1, exon junction complex associated factor |
| chr1_-_12898270 | 0.03 |
ENST00000235347.4
|
PRAMEF10
|
PRAME family member 10 |
| chr1_-_16237146 | 0.03 |
ENST00000375599.8
|
CPLANE2
|
ciliogenesis and planar polarity effector 2 |
| chr17_+_9825906 | 0.02 |
ENST00000262441.10
|
GLP2R
|
glucagon like peptide 2 receptor |
| chr3_+_49021071 | 0.02 |
ENST00000395458.6
|
NDUFAF3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr9_-_21995262 | 0.02 |
ENST00000494262.5
|
CDKN2A
|
cyclin dependent kinase inhibitor 2A |
| chr17_+_70104848 | 0.02 |
ENST00000392670.5
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr14_+_21797272 | 0.02 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr1_-_47190013 | 0.01 |
ENST00000294338.7
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr20_+_35968566 | 0.01 |
ENST00000373973.7
ENST00000349339.5 ENST00000489667.1 ENST00000538900.1 |
CNBD2
|
cyclic nucleotide binding domain containing 2 |
| chr7_+_21543020 | 0.01 |
ENST00000409508.8
ENST00000620169.4 ENST00000328843.10 |
DNAH11
|
dynein axonemal heavy chain 11 |
| chr19_-_5286163 | 0.01 |
ENST00000592099.5
ENST00000588012.5 ENST00000262963.10 ENST00000587303.5 ENST00000590509.5 |
PTPRS
|
protein tyrosine phosphatase receptor type S |
| chr6_-_138572523 | 0.01 |
ENST00000427025.6
|
NHSL1
|
NHS like 1 |
| chr8_-_109644766 | 0.00 |
ENST00000533065.5
ENST00000276646.14 |
SYBU
|
syntabulin |
| chr18_-_72865680 | 0.00 |
ENST00000397929.5
|
NETO1
|
neuropilin and tolloid like 1 |
| chr19_-_51034840 | 0.00 |
ENST00000529888.5
|
KLK12
|
kallikrein related peptidase 12 |
| chr19_-_51034892 | 0.00 |
ENST00000319590.8
ENST00000250351.4 |
KLK12
|
kallikrein related peptidase 12 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1902771 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.3 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.1 | 0.2 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.2 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) protein K27-linked deubiquitination(GO:1990167) |
| 0.0 | 0.2 | GO:0021816 | extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.7 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.3 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.8 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.6 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.7 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |