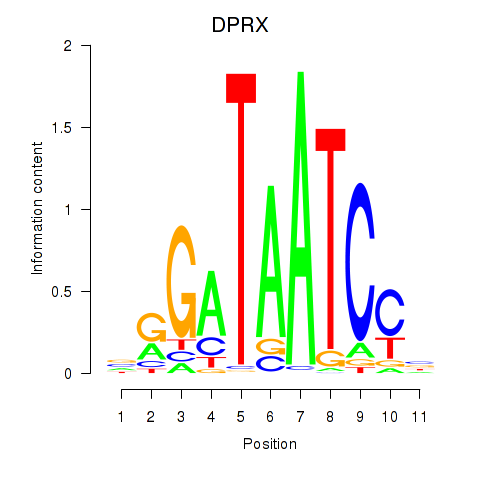
|
chr7_+_28412511
Show fit
|
0.76 |
ENST00000357727.7
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr8_+_103372388
Show fit
|
0.69 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr1_-_184974477
Show fit
|
0.65 |
ENST00000367511.4
|
NIBAN1
|
niban apoptosis regulator 1
|
|
chr1_-_13201409
Show fit
|
0.55 |
ENST00000625019.3
|
PRAMEF13
|
PRAME family member 13
|
|
chr1_-_153549120
Show fit
|
0.55 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3
|
|
chr6_+_26383176
Show fit
|
0.54 |
ENST00000416795.6
ENST00000494184.1
|
BTN2A2
|
butyrophilin subfamily 2 member A2
|
|
chr10_+_72692125
Show fit
|
0.53 |
ENST00000373053.7
ENST00000357157.10
|
MCU
|
mitochondrial calcium uniporter
|
|
chr11_-_126062782
Show fit
|
0.51 |
ENST00000531738.6
|
CDON
|
cell adhesion associated, oncogene regulated
|
|
chr1_-_173205543
Show fit
|
0.51 |
ENST00000367718.5
|
TNFSF4
|
TNF superfamily member 4
|
|
chr6_-_37257622
Show fit
|
0.49 |
ENST00000650812.1
ENST00000497775.1
ENST00000478262.2
ENST00000356757.7
|
ENSG00000286105.1
TMEM217
|
novel transmembrane protein
transmembrane protein 217
|
|
chr6_-_37257590
Show fit
|
0.49 |
ENST00000336655.7
|
TMEM217
|
transmembrane protein 217
|
|
chr6_-_37257643
Show fit
|
0.48 |
ENST00000651039.1
ENST00000652495.1
ENST00000652218.1
|
TMEM217
|
transmembrane protein 217
|
|
chr12_-_9760893
Show fit
|
0.44 |
ENST00000228434.7
ENST00000536709.1
|
CD69
|
CD69 molecule
|
|
chr9_-_22009272
Show fit
|
0.42 |
ENST00000380142.5
ENST00000276925.7
|
CDKN2B
|
cyclin dependent kinase inhibitor 2B
|
|
chr6_+_26383090
Show fit
|
0.42 |
ENST00000469230.5
ENST00000490025.5
ENST00000352867.6
ENST00000356709.9
ENST00000493275.5
ENST00000472507.5
ENST00000482536.5
ENST00000432533.6
ENST00000482842.1
|
BTN2A2
|
butyrophilin subfamily 2 member A2
|
|
chr6_+_4706133
Show fit
|
0.41 |
ENST00000328908.9
|
CDYL
|
chromodomain Y like
|
|
chr1_-_120054225
Show fit
|
0.40 |
ENST00000602566.6
|
NOTCH2
|
notch receptor 2
|
|
chr1_-_153549238
Show fit
|
0.38 |
ENST00000368713.8
|
S100A3
|
S100 calcium binding protein A3
|
|
chr1_+_182839338
Show fit
|
0.38 |
ENST00000367549.4
|
DHX9
|
DExH-box helicase 9
|
|
chr3_+_186996444
Show fit
|
0.37 |
ENST00000676633.1
|
ST6GAL1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1
|
|
chr1_+_21570303
Show fit
|
0.37 |
ENST00000374830.2
|
ALPL
|
alkaline phosphatase, biomineralization associated
|
|
chr16_-_58546702
Show fit
|
0.36 |
ENST00000567133.1
|
CNOT1
|
CCR4-NOT transcription complex subunit 1
|
|
chr22_-_42959852
Show fit
|
0.36 |
ENST00000402229.5
ENST00000407585.5
ENST00000453079.1
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr2_-_165953750
Show fit
|
0.35 |
ENST00000243344.8
ENST00000679799.1
ENST00000679840.1
ENST00000681606.1
ENST00000680448.1
|
TTC21B
|
tetratricopeptide repeat domain 21B
|
|
chr3_+_141386862
Show fit
|
0.33 |
ENST00000513258.5
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr17_-_15341577
Show fit
|
0.31 |
ENST00000543896.1
ENST00000395930.6
ENST00000539245.5
ENST00000539316.1
|
TEKT3
|
tektin 3
|
|
chr19_-_7040179
Show fit
|
0.31 |
ENST00000381394.9
|
MBD3L4
|
methyl-CpG binding domain protein 3 like 4
|
|
chr4_-_170003738
Show fit
|
0.30 |
ENST00000502832.1
ENST00000393704.3
|
MFAP3L
|
microfibril associated protein 3 like
|
|
chr9_+_37667997
Show fit
|
0.30 |
ENST00000539465.5
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chr12_+_93378625
Show fit
|
0.30 |
ENST00000546925.1
|
NUDT4
|
nudix hydrolase 4
|
|
chr12_+_78036248
Show fit
|
0.29 |
ENST00000644176.1
|
NAV3
|
neuron navigator 3
|
|
chr1_-_150808251
Show fit
|
0.28 |
ENST00000271651.8
ENST00000676970.1
ENST00000679260.1
ENST00000676751.1
ENST00000677887.1
|
CTSK
|
cathepsin K
|
|
chr17_+_79025612
Show fit
|
0.28 |
ENST00000392445.6
|
C1QTNF1
|
C1q and TNF related 1
|
|
chr3_+_111911604
Show fit
|
0.28 |
ENST00000495180.1
|
PHLDB2
|
pleckstrin homology like domain family B member 2
|
|
chr17_-_43942472
Show fit
|
0.26 |
ENST00000225992.8
|
PPY
|
pancreatic polypeptide
|
|
chr6_-_40587314
Show fit
|
0.26 |
ENST00000338305.7
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2
|
|
chr11_-_65862026
Show fit
|
0.26 |
ENST00000532134.5
|
CFL1
|
cofilin 1
|
|
chr2_-_49154433
Show fit
|
0.26 |
ENST00000454032.5
ENST00000304421.8
|
FSHR
|
follicle stimulating hormone receptor
|
|
chr1_+_179591876
Show fit
|
0.25 |
ENST00000294848.12
|
TDRD5
|
tudor domain containing 5
|
|
chr11_+_62190212
Show fit
|
0.25 |
ENST00000306238.3
|
SCGB1D1
|
secretoglobin family 1D member 1
|
|
chr2_-_49154507
Show fit
|
0.25 |
ENST00000406846.7
|
FSHR
|
follicle stimulating hormone receptor
|
|
chrX_+_154144242
Show fit
|
0.25 |
ENST00000369951.9
|
OPN1LW
|
opsin 1, long wave sensitive
|
|
chr1_+_179591819
Show fit
|
0.25 |
ENST00000444136.6
|
TDRD5
|
tudor domain containing 5
|
|
chrX_+_154182596
Show fit
|
0.23 |
ENST00000595290.6
|
OPN1MW
|
opsin 1, medium wave sensitive
|
|
chr5_-_67196791
Show fit
|
0.23 |
ENST00000256447.5
|
CD180
|
CD180 molecule
|
|
chr5_+_35856883
Show fit
|
0.23 |
ENST00000506850.5
ENST00000303115.8
ENST00000511982.1
|
IL7R
|
interleukin 7 receptor
|
|
chr6_-_33322803
Show fit
|
0.23 |
ENST00000446511.1
ENST00000446403.1
ENST00000374542.10
ENST00000266000.10
ENST00000414083.6
ENST00000620164.4
|
DAXX
|
death domain associated protein
|
|
chr7_-_92833896
Show fit
|
0.22 |
ENST00000265734.8
|
CDK6
|
cyclin dependent kinase 6
|
|
chr1_+_156147350
Show fit
|
0.22 |
ENST00000435124.5
ENST00000633494.1
|
SEMA4A
|
semaphorin 4A
|
|
chr10_+_102132994
Show fit
|
0.22 |
ENST00000413464.6
ENST00000278070.7
|
PPRC1
|
PPARG related coactivator 1
|
|
chr5_+_140966466
Show fit
|
0.22 |
ENST00000615316.1
ENST00000289269.7
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr8_+_53851786
Show fit
|
0.22 |
ENST00000297313.8
ENST00000344277.10
|
RGS20
|
regulator of G protein signaling 20
|
|
chr1_-_13347134
Show fit
|
0.21 |
ENST00000334600.7
|
PRAMEF14
|
PRAME family member 14
|
|
chr10_+_95755737
Show fit
|
0.21 |
ENST00000543964.6
|
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1
|
|
chr10_+_95755652
Show fit
|
0.21 |
ENST00000371207.8
|
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1
|
|
chr19_-_12156710
Show fit
|
0.21 |
ENST00000455799.1
ENST00000439556.3
|
ZNF625
|
zinc finger protein 625
|
|
chr7_+_134891566
Show fit
|
0.21 |
ENST00000424922.5
ENST00000495522.1
|
CALD1
|
caldesmon 1
|
|
chr11_-_96389936
Show fit
|
0.21 |
ENST00000680763.1
ENST00000679856.1
ENST00000643839.1
ENST00000645500.1
ENST00000530106.2
|
CCDC82
|
coiled-coil domain containing 82
|
|
chr1_+_50103903
Show fit
|
0.20 |
ENST00000371827.5
|
ELAVL4
|
ELAV like RNA binding protein 4
|
|
chr9_+_113463697
Show fit
|
0.20 |
ENST00000317613.10
|
RGS3
|
regulator of G protein signaling 3
|
|
chr12_-_113136224
Show fit
|
0.20 |
ENST00000546530.5
ENST00000261729.9
|
RASAL1
|
RAS protein activator like 1
|
|
chr19_-_58558871
Show fit
|
0.20 |
ENST00000595957.5
|
UBE2M
|
ubiquitin conjugating enzyme E2 M
|
|
chr22_-_31107517
Show fit
|
0.20 |
ENST00000400299.6
ENST00000611680.1
|
SELENOM
|
selenoprotein M
|
|
chr4_-_103077282
Show fit
|
0.20 |
ENST00000503230.5
ENST00000503818.1
|
SLC9B2
|
solute carrier family 9 member B2
|
|
chr21_+_43728851
Show fit
|
0.19 |
ENST00000327574.4
|
PDXK
|
pyridoxal kinase
|
|
chr6_+_42050876
Show fit
|
0.19 |
ENST00000465926.5
ENST00000482432.1
|
TAF8
|
TATA-box binding protein associated factor 8
|
|
chr3_-_112845950
Show fit
|
0.19 |
ENST00000398214.5
|
CD200R1L
|
CD200 receptor 1 like
|
|
chr14_-_100568070
Show fit
|
0.19 |
ENST00000557378.6
ENST00000443071.6
ENST00000637646.1
|
BEGAIN
|
brain enriched guanylate kinase associated
|
|
chr11_-_66907891
Show fit
|
0.19 |
ENST00000393955.6
|
PC
|
pyruvate carboxylase
|
|
chr6_+_37257762
Show fit
|
0.19 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family member 22B
|
|
chr1_+_224183262
Show fit
|
0.19 |
ENST00000391877.3
|
DEGS1
|
delta 4-desaturase, sphingolipid 1
|
|
chr17_-_35119733
Show fit
|
0.18 |
ENST00000460118.6
ENST00000335858.11
|
RAD51D
|
RAD51 paralog D
|
|
chr11_-_105035113
Show fit
|
0.18 |
ENST00000526568.5
ENST00000531166.5
ENST00000534497.5
ENST00000527979.5
ENST00000533400.6
ENST00000528974.1
ENST00000525825.5
ENST00000353247.9
ENST00000446369.5
ENST00000436863.7
|
CASP1
|
caspase 1
|
|
chr3_+_98353854
Show fit
|
0.18 |
ENST00000354924.2
|
OR5K4
|
olfactory receptor family 5 subfamily K member 4
|
|
chr12_+_132986359
Show fit
|
0.18 |
ENST00000328654.10
|
ZNF26
|
zinc finger protein 26
|
|
chr16_+_67528799
Show fit
|
0.18 |
ENST00000379312.7
ENST00000042381.9
ENST00000540839.7
|
RIPOR1
|
RHO family interacting cell polarization regulator 1
|
|
chr22_-_30471986
Show fit
|
0.18 |
ENST00000401751.5
ENST00000402286.5
ENST00000403066.5
ENST00000215812.9
|
SEC14L3
|
SEC14 like lipid binding 3
|
|
chr12_-_101830926
Show fit
|
0.17 |
ENST00000299314.12
|
GNPTAB
|
N-acetylglucosamine-1-phosphate transferase subunits alpha and beta
|
|
chr17_-_39225936
Show fit
|
0.17 |
ENST00000333461.6
|
STAC2
|
SH3 and cysteine rich domain 2
|
|
chr9_+_2110354
Show fit
|
0.17 |
ENST00000634772.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr19_-_12723925
Show fit
|
0.17 |
ENST00000425528.6
ENST00000589337.5
ENST00000588216.5
|
TNPO2
|
transportin 2
|
|
chr5_-_124745315
Show fit
|
0.17 |
ENST00000306315.9
|
ZNF608
|
zinc finger protein 608
|
|
chr1_+_200894892
Show fit
|
0.17 |
ENST00000413687.3
|
INAVA
|
innate immunity activator
|
|
chrX_+_15507302
Show fit
|
0.17 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr21_+_42653585
Show fit
|
0.16 |
ENST00000291539.11
|
PDE9A
|
phosphodiesterase 9A
|
|
chr1_+_236394268
Show fit
|
0.16 |
ENST00000334232.9
|
EDARADD
|
EDAR associated death domain
|
|
chr3_+_63967738
Show fit
|
0.16 |
ENST00000484332.1
|
ATXN7
|
ataxin 7
|
|
chr21_-_33643926
Show fit
|
0.16 |
ENST00000438788.1
|
CRYZL1
|
crystallin zeta like 1
|
|
chr14_-_100568475
Show fit
|
0.16 |
ENST00000553553.6
|
BEGAIN
|
brain enriched guanylate kinase associated
|
|
chr7_+_134779625
Show fit
|
0.16 |
ENST00000454108.5
ENST00000361675.7
|
CALD1
|
caldesmon 1
|
|
chr12_-_76878985
Show fit
|
0.16 |
ENST00000547435.1
ENST00000552330.5
ENST00000311083.10
ENST00000546966.5
|
CSRP2
|
cysteine and glycine rich protein 2
|
|
chr14_-_20587962
Show fit
|
0.16 |
ENST00000557105.5
ENST00000398008.6
ENST00000620912.4
ENST00000555841.5
ENST00000443456.6
ENST00000432835.6
ENST00000557503.5
ENST00000398009.6
ENST00000554842.1
|
RNASE11
|
ribonuclease A family member 11 (inactive)
|
|
chr14_-_20587716
Show fit
|
0.15 |
ENST00000553849.1
|
RNASE11
|
ribonuclease A family member 11 (inactive)
|
|
chr20_+_142573
Show fit
|
0.15 |
ENST00000382398.4
|
DEFB126
|
defensin beta 126
|
|
chr8_-_30812867
Show fit
|
0.15 |
ENST00000518243.5
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta
|
|
chr19_+_7049321
Show fit
|
0.15 |
ENST00000381393.3
|
MBD3L2
|
methyl-CpG binding domain protein 3 like 2
|
|
chr19_+_7030578
Show fit
|
0.15 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3 like 5
|
|
chr4_-_151227881
Show fit
|
0.15 |
ENST00000652233.1
ENST00000514152.5
|
SH3D19
|
SH3 domain containing 19
|
|
chr7_+_134779663
Show fit
|
0.15 |
ENST00000361901.6
|
CALD1
|
caldesmon 1
|
|
chr5_+_141370236
Show fit
|
0.15 |
ENST00000576222.2
ENST00000618934.1
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr17_-_35119801
Show fit
|
0.15 |
ENST00000592577.5
ENST00000590016.5
ENST00000345365.11
|
RAD51D
|
RAD51 paralog D
|
|
chr1_+_15659869
Show fit
|
0.14 |
ENST00000345034.1
|
RSC1A1
|
regulator of solute carriers 1
|
|
chr3_-_42875871
Show fit
|
0.14 |
ENST00000316161.6
ENST00000437102.1
|
CYP8B1
|
cytochrome P450 family 8 subfamily B member 1
|
|
chr1_+_159439722
Show fit
|
0.14 |
ENST00000641630.1
ENST00000423932.6
|
OR10J1
|
olfactory receptor family 10 subfamily J member 1
|
|
chr3_-_88149815
Show fit
|
0.14 |
ENST00000467332.1
ENST00000462901.5
|
CGGBP1
|
CGG triplet repeat binding protein 1
|
|
chr11_-_5269933
Show fit
|
0.14 |
ENST00000396895.3
|
HBE1
|
hemoglobin subunit epsilon 1
|
|
chr17_-_4739866
Show fit
|
0.14 |
ENST00000574412.6
ENST00000293778.12
|
CXCL16
|
C-X-C motif chemokine ligand 16
|
|
chr1_+_16440700
Show fit
|
0.14 |
ENST00000504551.6
ENST00000457722.6
ENST00000337132.10
ENST00000443980.6
|
NECAP2
|
NECAP endocytosis associated 2
|
|
chr19_+_35358460
Show fit
|
0.14 |
ENST00000327809.5
|
FFAR3
|
free fatty acid receptor 3
|
|
chr7_+_134891400
Show fit
|
0.14 |
ENST00000393118.6
|
CALD1
|
caldesmon 1
|
|
chr12_+_103965835
Show fit
|
0.14 |
ENST00000266775.13
ENST00000544861.5
|
TDG
|
thymine DNA glycosylase
|
|
chrX_+_53422890
Show fit
|
0.13 |
ENST00000457095.5
|
RIBC1
|
RIB43A domain with coiled-coils 1
|
|
chr4_-_103076688
Show fit
|
0.13 |
ENST00000394785.9
|
SLC9B2
|
solute carrier family 9 member B2
|
|
chr12_+_103965798
Show fit
|
0.13 |
ENST00000436021.6
|
TDG
|
thymine DNA glycosylase
|
|
chr10_-_101818405
Show fit
|
0.13 |
ENST00000357797.9
ENST00000370094.7
|
OGA
|
O-GlcNAcase
|
|
chr1_+_13892791
Show fit
|
0.13 |
ENST00000636564.1
ENST00000636203.1
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr16_+_21233672
Show fit
|
0.13 |
ENST00000311620.7
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B
|
|
chr19_-_7021431
Show fit
|
0.13 |
ENST00000636986.2
ENST00000637800.1
|
MBD3L2B
|
methyl-CpG binding domain protein 3 like 2B
|
|
chr2_-_202870862
Show fit
|
0.13 |
ENST00000454326.5
ENST00000432273.5
ENST00000450143.5
ENST00000411681.5
|
ICA1L
|
islet cell autoantigen 1 like
|
|
chr8_+_49909783
Show fit
|
0.12 |
ENST00000518864.5
|
SNTG1
|
syntrophin gamma 1
|
|
chr8_-_30812773
Show fit
|
0.12 |
ENST00000221138.9
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta
|
|
chr1_-_246931892
Show fit
|
0.12 |
ENST00000648844.2
|
AHCTF1
|
AT-hook containing transcription factor 1
|
|
chr7_+_66114814
Show fit
|
0.11 |
ENST00000431089.6
ENST00000398684.6
ENST00000395326.8
ENST00000338592.5
|
CRCP
|
CGRP receptor component
|
|
chr19_+_50025714
Show fit
|
0.11 |
ENST00000598809.5
ENST00000595661.5
ENST00000391821.6
|
ZNF473
|
zinc finger protein 473
|
|
chr9_+_4839761
Show fit
|
0.11 |
ENST00000448872.6
ENST00000441844.2
|
RCL1
|
RNA terminal phosphate cyclase like 1
|
|
chr17_-_28897724
Show fit
|
0.11 |
ENST00000394908.9
|
FLOT2
|
flotillin 2
|
|
chr17_-_28897602
Show fit
|
0.11 |
ENST00000394906.6
ENST00000585169.5
|
FLOT2
|
flotillin 2
|
|
chr12_+_103965863
Show fit
|
0.11 |
ENST00000392872.8
ENST00000537100.5
|
TDG
|
thymine DNA glycosylase
|
|
chr19_+_55654115
Show fit
|
0.11 |
ENST00000450554.6
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2
|
|
chr1_+_28668746
Show fit
|
0.11 |
ENST00000361872.8
ENST00000294409.2
ENST00000373816.6
|
GMEB1
|
glucocorticoid modulatory element binding protein 1
|
|
chrX_-_53422644
Show fit
|
0.11 |
ENST00000322213.9
ENST00000375340.10
ENST00000674590.1
|
SMC1A
|
structural maintenance of chromosomes 1A
|
|
chr9_+_113444725
Show fit
|
0.11 |
ENST00000374140.6
|
RGS3
|
regulator of G protein signaling 3
|
|
chr1_-_170074568
Show fit
|
0.10 |
ENST00000367767.5
ENST00000361580.7
ENST00000538366.5
|
KIFAP3
|
kinesin associated protein 3
|
|
chr11_-_96389857
Show fit
|
0.10 |
ENST00000680532.1
ENST00000681014.1
ENST00000679708.1
ENST00000645439.1
ENST00000645366.1
ENST00000680728.1
ENST00000680859.1
ENST00000679788.1
ENST00000681164.1
ENST00000646818.2
ENST00000680171.1
ENST00000679696.1
ENST00000681200.1
ENST00000679960.1
ENST00000647080.1
ENST00000646050.1
ENST00000681451.1
ENST00000644686.1
ENST00000680052.1
ENST00000679616.1
ENST00000530203.2
|
CCDC82
|
coiled-coil domain containing 82
|
|
chr14_+_21797272
Show fit
|
0.10 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1
|
|
chr6_-_22297028
Show fit
|
0.10 |
ENST00000306482.2
|
PRL
|
prolactin
|
|
chr5_-_178590367
Show fit
|
0.10 |
ENST00000390654.8
|
COL23A1
|
collagen type XXIII alpha 1 chain
|
|
chr8_-_70608407
Show fit
|
0.10 |
ENST00000262213.7
|
TRAM1
|
translocation associated membrane protein 1
|
|
chr17_-_67996428
Show fit
|
0.10 |
ENST00000580729.3
|
C17orf58
|
chromosome 17 open reading frame 58
|
|
chr1_+_20070156
Show fit
|
0.10 |
ENST00000375108.4
|
PLA2G5
|
phospholipase A2 group V
|
|
chr2_-_25252251
Show fit
|
0.10 |
ENST00000380746.8
ENST00000402667.1
|
DNMT3A
|
DNA methyltransferase 3 alpha
|
|
chr1_+_12773738
Show fit
|
0.10 |
ENST00000357726.5
|
PRAMEF12
|
PRAME family member 12
|
|
chr14_+_76826372
Show fit
|
0.09 |
ENST00000393774.7
ENST00000555189.1
|
LRRC74A
|
leucine rich repeat containing 74A
|
|
chr2_+_233307806
Show fit
|
0.09 |
ENST00000447536.5
ENST00000409110.6
|
SAG
|
S-antigen visual arrestin
|
|
chr10_-_13972355
Show fit
|
0.09 |
ENST00000264546.10
|
FRMD4A
|
FERM domain containing 4A
|
|
chr1_+_74235377
Show fit
|
0.09 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr2_-_25252072
Show fit
|
0.09 |
ENST00000683760.1
|
DNMT3A
|
DNA methyltransferase 3 alpha
|
|
chr4_+_152779934
Show fit
|
0.08 |
ENST00000451320.6
ENST00000429148.6
ENST00000353617.7
ENST00000405727.6
ENST00000356064.3
|
ARFIP1
|
ADP ribosylation factor interacting protein 1
|
|
chr9_+_17135017
Show fit
|
0.08 |
ENST00000380641.4
ENST00000380647.8
|
CNTLN
|
centlein
|
|
chr2_-_157327699
Show fit
|
0.08 |
ENST00000397283.6
|
ERMN
|
ermin
|
|
chr12_-_110499546
Show fit
|
0.08 |
ENST00000552130.6
|
VPS29
|
VPS29 retromer complex component
|
|
chr5_-_62403506
Show fit
|
0.08 |
ENST00000680062.1
|
DIMT1
|
DIMT1 rRNA methyltransferase and ribosome maturation factor
|
|
chr4_-_165279679
Show fit
|
0.08 |
ENST00000505354.2
|
GK3P
|
glycerol kinase 3 pseudogene
|
|
chr22_-_19150292
Show fit
|
0.08 |
ENST00000086933.3
|
GSC2
|
goosecoid homeobox 2
|
|
chr3_+_36993798
Show fit
|
0.08 |
ENST00000455445.6
ENST00000539477.6
ENST00000435176.5
ENST00000429117.5
ENST00000441265.6
|
MLH1
|
mutL homolog 1
|
|
chr22_-_26512377
Show fit
|
0.07 |
ENST00000407431.5
ENST00000455080.5
ENST00000407690.6
ENST00000418876.5
ENST00000440258.1
ENST00000407148.5
ENST00000420242.5
ENST00000405938.5
|
TFIP11
|
tuftelin interacting protein 11
|
|
chr8_-_123025627
Show fit
|
0.07 |
ENST00000519018.5
|
DERL1
|
derlin 1
|
|
chr1_+_119507203
Show fit
|
0.07 |
ENST00000369413.8
ENST00000528909.1
|
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1
|
|
chr10_-_70602687
Show fit
|
0.07 |
ENST00000638674.1
|
PRF1
|
perforin 1
|
|
chr2_-_127858107
Show fit
|
0.07 |
ENST00000409955.1
ENST00000272645.9
|
POLR2D
|
RNA polymerase II subunit D
|
|
chr7_-_135728177
Show fit
|
0.07 |
ENST00000682651.1
ENST00000354042.8
|
SLC13A4
|
solute carrier family 13 member 4
|
|
chr14_-_106557465
Show fit
|
0.07 |
ENST00000390625.3
|
IGHV3-49
|
immunoglobulin heavy variable 3-49
|
|
chr18_-_3845292
Show fit
|
0.07 |
ENST00000400145.6
|
DLGAP1
|
DLG associated protein 1
|
|
chr9_-_107489754
Show fit
|
0.07 |
ENST00000610832.1
ENST00000374672.5
|
KLF4
|
Kruppel like factor 4
|
|
chr20_+_31819348
Show fit
|
0.07 |
ENST00000375985.5
|
MYLK2
|
myosin light chain kinase 2
|
|
chr4_-_122621011
Show fit
|
0.07 |
ENST00000611104.2
ENST00000648588.1
|
IL21
|
interleukin 21
|
|
chr8_-_144475808
Show fit
|
0.07 |
ENST00000377317.5
|
FOXH1
|
forkhead box H1
|
|
chr5_-_59768631
Show fit
|
0.07 |
ENST00000502575.1
ENST00000507116.5
|
PDE4D
|
phosphodiesterase 4D
|
|
chrX_-_53422170
Show fit
|
0.07 |
ENST00000675504.1
|
SMC1A
|
structural maintenance of chromosomes 1A
|
|
chr20_+_10218808
Show fit
|
0.07 |
ENST00000254976.7
ENST00000304886.6
|
SNAP25
|
synaptosome associated protein 25
|
|
chr10_-_70602759
Show fit
|
0.07 |
ENST00000373209.2
|
PRF1
|
perforin 1
|
|
chr8_+_47260922
Show fit
|
0.06 |
ENST00000518074.5
ENST00000297423.9
ENST00000524006.5
|
SPIDR
|
scaffold protein involved in DNA repair
|
|
chr20_+_31819302
Show fit
|
0.06 |
ENST00000375994.6
|
MYLK2
|
myosin light chain kinase 2
|
|
chr10_-_70602731
Show fit
|
0.06 |
ENST00000441259.2
|
PRF1
|
perforin 1
|
|
chr3_+_36993764
Show fit
|
0.06 |
ENST00000674019.1
ENST00000458205.6
|
MLH1
|
mutL homolog 1
|
|
chr17_-_50707855
Show fit
|
0.06 |
ENST00000285243.7
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr12_+_53954870
Show fit
|
0.06 |
ENST00000243103.4
|
HOXC12
|
homeobox C12
|
|
chr1_-_13155961
Show fit
|
0.06 |
ENST00000624207.1
|
PRAMEF26
|
PRAME family member 26
|
|
chr3_+_188947199
Show fit
|
0.06 |
ENST00000433971.5
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chr3_+_130931893
Show fit
|
0.06 |
ENST00000504612.5
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1
|
|
chr15_-_52569197
Show fit
|
0.06 |
ENST00000563277.5
ENST00000566423.5
|
ARPP19
|
cAMP regulated phosphoprotein 19
|
|
chr1_+_42682388
Show fit
|
0.06 |
ENST00000321358.12
ENST00000332220.10
|
YBX1
|
Y-box binding protein 1
|
|
chr15_-_52569126
Show fit
|
0.06 |
ENST00000569723.5
ENST00000567669.5
ENST00000569281.6
ENST00000563566.5
ENST00000567830.1
|
ARPP19
|
cAMP regulated phosphoprotein 19
|
|
chr1_+_13171848
Show fit
|
0.06 |
ENST00000415919.3
|
PRAMEF9
|
PRAME family member 9
|
|
chr19_+_8413270
Show fit
|
0.06 |
ENST00000381035.8
ENST00000595142.5
ENST00000601724.5
ENST00000601283.5
ENST00000215555.7
ENST00000595213.1
|
MARCHF2
|
membrane associated ring-CH-type finger 2
|
|
chr2_+_68974573
Show fit
|
0.06 |
ENST00000673932.3
ENST00000377938.4
|
GKN1
|
gastrokine 1
|
|
chr12_+_93378550
Show fit
|
0.06 |
ENST00000550056.5
ENST00000549992.5
ENST00000548662.5
ENST00000547014.5
|
NUDT4
|
nudix hydrolase 4
|
|
chr18_+_58862904
Show fit
|
0.06 |
ENST00000591083.5
|
ZNF532
|
zinc finger protein 532
|
|
chr2_-_29921580
Show fit
|
0.06 |
ENST00000389048.8
|
ALK
|
ALK receptor tyrosine kinase
|
|
chr8_+_47260878
Show fit
|
0.06 |
ENST00000541342.2
|
SPIDR
|
scaffold protein involved in DNA repair
|
|
chr20_-_45308297
Show fit
|
0.05 |
ENST00000353917.10
ENST00000360607.10
|
MATN4
|
matrilin 4
|
|
chr3_+_49021071
Show fit
|
0.05 |
ENST00000395458.6
|
NDUFAF3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3
|
|
chr1_+_59297057
Show fit
|
0.05 |
ENST00000303721.12
|
FGGY
|
FGGY carbohydrate kinase domain containing
|
|
chr20_+_1895365
Show fit
|
0.05 |
ENST00000358771.5
|
SIRPA
|
signal regulatory protein alpha
|
|
chr8_-_109644766
Show fit
|
0.05 |
ENST00000533065.5
ENST00000276646.14
|
SYBU
|
syntabulin
|
|
chr18_-_49849827
Show fit
|
0.05 |
ENST00000592688.1
|
MYO5B
|
myosin VB
|
|
chr6_+_29395631
Show fit
|
0.05 |
ENST00000642051.1
|
OR12D2
|
olfactory receptor family 12 subfamily D member 2
|
|
chr17_+_50834581
Show fit
|
0.05 |
ENST00000426127.1
|
WFIKKN2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2
|
|
chr19_-_50025936
Show fit
|
0.05 |
ENST00000596445.5
ENST00000599538.5
|
VRK3
|
VRK serine/threonine kinase 3
|
|
chr4_-_152779710
Show fit
|
0.05 |
ENST00000304337.3
|
TIGD4
|
tigger transposable element derived 4
|