Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for DRGX_PROP1
Z-value: 0.64
Transcription factors associated with DRGX_PROP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
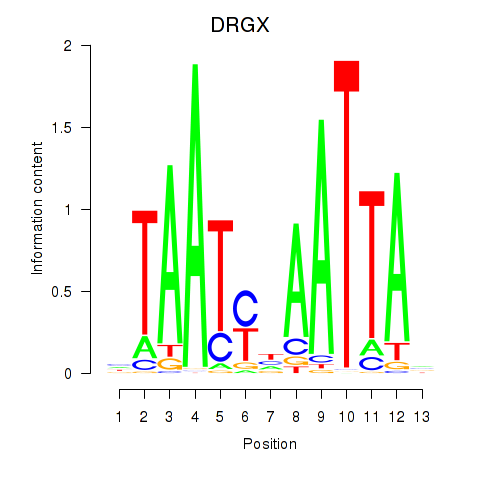
DRGX
|
ENSG00000165606.10 | DRGX |
|
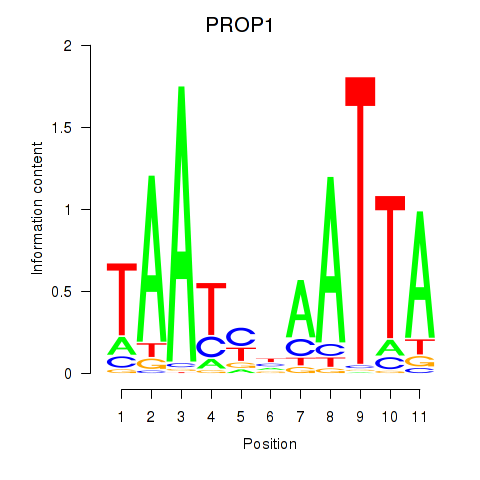
PROP1
|
ENSG00000175325.2 | PROP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PROP1 | hg38_v1_chr5_-_177996242_177996242 | 0.15 | 4.9e-01 | Click! |
Activity profile of DRGX_PROP1 motif
Sorted Z-values of DRGX_PROP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of DRGX_PROP1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_91906280 | 4.75 |
ENST00000370399.6
|
TGFBR3
|
transforming growth factor beta receptor 3 |
| chr1_+_84181630 | 3.06 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_-_55269038 | 2.38 |
ENST00000417363.5
ENST00000412530.1 ENST00000366137.6 ENST00000420637.5 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr9_-_123184233 | 1.88 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr6_-_75363003 | 1.83 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr3_-_18438767 | 1.67 |
ENST00000454909.6
|
SATB1
|
SATB homeobox 1 |
| chr6_+_121437378 | 1.66 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr8_-_92095215 | 1.53 |
ENST00000360348.6
ENST00000520428.5 ENST00000518992.5 ENST00000520556.5 ENST00000518317.5 ENST00000521319.5 ENST00000521375.5 ENST00000518449.5 ENST00000613886.4 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr16_+_68085344 | 1.52 |
ENST00000575270.5
ENST00000346183.8 |
NFATC3
|
nuclear factor of activated T cells 3 |
| chr3_-_125055987 | 1.44 |
ENST00000311127.9
|
HEG1
|
heart development protein with EGF like domains 1 |
| chr16_+_68085420 | 1.43 |
ENST00000349223.9
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr16_+_68085552 | 1.28 |
ENST00000329524.8
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr1_+_84164962 | 1.26 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr6_+_116369837 | 1.24 |
ENST00000645988.1
|
DSE
|
dermatan sulfate epimerase |
| chr2_-_182427014 | 1.08 |
ENST00000409365.5
ENST00000351439.9 |
PDE1A
|
phosphodiesterase 1A |
| chr3_-_27722699 | 1.06 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr7_-_120858066 | 1.04 |
ENST00000222747.8
|
TSPAN12
|
tetraspanin 12 |
| chr5_+_90640718 | 1.04 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr2_+_178320228 | 1.02 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr13_+_53028806 | 1.00 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr10_+_68106109 | 0.98 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr16_-_67483541 | 0.96 |
ENST00000290953.3
|
AGRP
|
agouti related neuropeptide |
| chr12_+_19205294 | 0.96 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr14_-_75069478 | 0.93 |
ENST00000555463.1
|
ACYP1
|
acylphosphatase 1 |
| chr6_+_30914205 | 0.93 |
ENST00000672801.1
ENST00000321897.9 ENST00000625423.2 ENST00000676266.1 ENST00000428017.5 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr6_+_30914329 | 0.89 |
ENST00000541562.6
|
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr5_-_111976925 | 0.83 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr6_-_132659178 | 0.81 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr2_-_55334529 | 0.81 |
ENST00000645860.1
ENST00000642563.1 ENST00000647396.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr9_+_2159672 | 0.81 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr4_+_155666718 | 0.77 |
ENST00000621234.4
ENST00000511108.5 |
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr8_-_85341659 | 0.76 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr10_-_49188380 | 0.75 |
ENST00000374153.7
ENST00000374148.1 ENST00000374151.7 |
TMEM273
|
transmembrane protein 273 |
| chr1_-_93681829 | 0.74 |
ENST00000260502.11
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr1_+_243256034 | 0.74 |
ENST00000366541.8
|
SDCCAG8
|
SHH signaling and ciliogenesis regulator SDCCAG8 |
| chr18_-_55322215 | 0.71 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr15_+_48191648 | 0.70 |
ENST00000646012.1
ENST00000561127.5 ENST00000647546.1 ENST00000559641.5 ENST00000417307.3 |
SLC12A1
CTXN2
|
solute carrier family 12 member 1 cortexin 2 |
| chr15_-_52678560 | 0.70 |
ENST00000562351.2
ENST00000261844.11 ENST00000399202.8 ENST00000562135.5 |
FAM214A
|
family with sequence similarity 214 member A |
| chrX_+_77910656 | 0.68 |
ENST00000343533.9
ENST00000341514.11 ENST00000645454.1 ENST00000642651.1 ENST00000644362.1 |
ATP7A
PGK1
|
ATPase copper transporting alpha phosphoglycerate kinase 1 |
| chr6_-_111483700 | 0.68 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr1_+_14929734 | 0.68 |
ENST00000376028.8
ENST00000400798.6 |
KAZN
|
kazrin, periplakin interacting protein |
| chr1_+_248508073 | 0.68 |
ENST00000641804.1
|
OR2G6
|
olfactory receptor family 2 subfamily G member 6 |
| chr16_+_24537693 | 0.68 |
ENST00000564314.5
ENST00000567686.5 |
RBBP6
|
RB binding protein 6, ubiquitin ligase |
| chr15_+_57219411 | 0.66 |
ENST00000543579.5
ENST00000537840.5 ENST00000343827.7 |
TCF12
|
transcription factor 12 |
| chr8_+_26293112 | 0.64 |
ENST00000523925.5
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr1_+_77918128 | 0.63 |
ENST00000342754.5
|
NEXN
|
nexilin F-actin binding protein |
| chr2_-_58241259 | 0.63 |
ENST00000481670.2
ENST00000403676.5 ENST00000427708.6 ENST00000403295.7 ENST00000233741.9 ENST00000446381.5 ENST00000417361.1 ENST00000402135.7 ENST00000449070.5 |
FANCL
|
FA complementation group L |
| chr18_-_55321986 | 0.62 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr1_-_21050952 | 0.60 |
ENST00000264211.12
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma 3 |
| chr11_-_118252279 | 0.59 |
ENST00000525386.5
ENST00000527472.1 ENST00000278949.9 |
MPZL3
|
myelin protein zero like 3 |
| chr9_+_2159850 | 0.58 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_-_218010202 | 0.56 |
ENST00000646520.1
|
TNS1
|
tensin 1 |
| chr5_-_135954962 | 0.56 |
ENST00000522943.5
ENST00000514447.2 ENST00000274507.6 |
LECT2
|
leukocyte cell derived chemotaxin 2 |
| chr5_-_55173173 | 0.54 |
ENST00000296733.5
ENST00000322374.10 ENST00000381375.7 |
CDC20B
|
cell division cycle 20B |
| chr12_-_118359639 | 0.53 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr4_-_99435396 | 0.49 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr17_+_46511511 | 0.49 |
ENST00000576629.5
|
LRRC37A2
|
leucine rich repeat containing 37 member A2 |
| chr5_+_141192330 | 0.49 |
ENST00000239446.6
|
PCDHB10
|
protocadherin beta 10 |
| chr8_+_12104389 | 0.49 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D |
| chr8_-_17895487 | 0.48 |
ENST00000427924.5
ENST00000381841.4 |
FGL1
|
fibrinogen like 1 |
| chr5_+_36606355 | 0.48 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr12_-_7503744 | 0.47 |
ENST00000396620.7
ENST00000432237.3 |
CD163
|
CD163 molecule |
| chr3_-_27722316 | 0.46 |
ENST00000449599.4
|
EOMES
|
eomesodermin |
| chr9_+_74615582 | 0.46 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B |
| chr8_-_17895403 | 0.45 |
ENST00000381840.5
ENST00000398054.5 |
FGL1
|
fibrinogen like 1 |
| chr10_+_35175586 | 0.45 |
ENST00000494479.5
ENST00000463314.5 ENST00000342105.7 ENST00000495301.1 |
CREM
|
cAMP responsive element modulator |
| chr3_-_197573323 | 0.44 |
ENST00000358186.6
ENST00000431056.5 |
BDH1
|
3-hydroxybutyrate dehydrogenase 1 |
| chr19_-_57974527 | 0.44 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr7_+_107583919 | 0.44 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr4_-_99435336 | 0.43 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr15_+_75336018 | 0.41 |
ENST00000567195.5
|
COMMD4
|
COMM domain containing 4 |
| chr2_+_233691607 | 0.39 |
ENST00000373424.5
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase family 1 member A6 |
| chrX_+_120604084 | 0.38 |
ENST00000371317.10
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr5_-_56116946 | 0.38 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr22_-_28711931 | 0.37 |
ENST00000434810.5
ENST00000456369.5 |
CHEK2
|
checkpoint kinase 2 |
| chrX_-_19799751 | 0.36 |
ENST00000379698.8
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr3_+_111674654 | 0.36 |
ENST00000636933.1
ENST00000393934.7 ENST00000477665.2 |
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2 |
| chr16_-_69339493 | 0.36 |
ENST00000562595.5
ENST00000615447.1 ENST00000306875.10 ENST00000562081.2 |
COG8
|
component of oligomeric golgi complex 8 |
| chr12_-_7503841 | 0.35 |
ENST00000359156.8
|
CD163
|
CD163 molecule |
| chr9_+_133636355 | 0.35 |
ENST00000393056.8
|
DBH
|
dopamine beta-hydroxylase |
| chr9_+_72577369 | 0.34 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr8_-_56211257 | 0.34 |
ENST00000316981.8
ENST00000423799.6 ENST00000429357.2 |
PLAG1
|
PLAG1 zinc finger |
| chr3_-_165837412 | 0.34 |
ENST00000479451.5
ENST00000488954.1 ENST00000264381.8 |
BCHE
|
butyrylcholinesterase |
| chr4_+_40196907 | 0.33 |
ENST00000622175.4
ENST00000619474.4 ENST00000615083.4 ENST00000610353.4 ENST00000614836.1 |
RHOH
|
ras homolog family member H |
| chr5_-_111758061 | 0.33 |
ENST00000509979.5
ENST00000513100.5 ENST00000508161.5 ENST00000455559.6 |
NREP
|
neuronal regeneration related protein |
| chr6_-_41071825 | 0.32 |
ENST00000468811.5
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr22_-_28712136 | 0.31 |
ENST00000464581.6
|
CHEK2
|
checkpoint kinase 2 |
| chr4_+_40197023 | 0.30 |
ENST00000381799.10
|
RHOH
|
ras homolog family member H |
| chr6_+_26087417 | 0.30 |
ENST00000357618.10
ENST00000309234.10 |
HFE
|
homeostatic iron regulator |
| chr20_+_33562306 | 0.30 |
ENST00000344201.7
|
CBFA2T2
|
CBFA2/RUNX1 partner transcriptional co-repressor 2 |
| chr3_+_111978996 | 0.29 |
ENST00000273359.8
ENST00000494817.1 |
ABHD10
|
abhydrolase domain containing 10, depalmitoylase |
| chr7_-_120857124 | 0.28 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chr15_+_45252228 | 0.28 |
ENST00000560438.5
ENST00000347644.8 |
SLC28A2
|
solute carrier family 28 member 2 |
| chr2_-_201739205 | 0.27 |
ENST00000681558.1
ENST00000681495.1 |
ALS2
|
alsin Rho guanine nucleotide exchange factor ALS2 |
| chr20_+_33235987 | 0.27 |
ENST00000375422.6
ENST00000375413.8 ENST00000354297.9 |
BPIFA1
|
BPI fold containing family A member 1 |
| chr2_+_108621260 | 0.26 |
ENST00000409441.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr11_+_55262152 | 0.26 |
ENST00000417545.5
|
TRIM48
|
tripartite motif containing 48 |
| chr1_+_244051275 | 0.25 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr22_-_28306645 | 0.25 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr11_+_108116688 | 0.25 |
ENST00000672284.1
|
ACAT1
|
acetyl-CoA acetyltransferase 1 |
| chr19_+_55769118 | 0.25 |
ENST00000341750.5
|
RFPL4AL1
|
ret finger protein like 4A like 1 |
| chr15_-_56243829 | 0.24 |
ENST00000559447.8
ENST00000673997.1 |
RFX7
|
regulatory factor X7 |
| chr5_-_138139382 | 0.24 |
ENST00000265191.4
|
NME5
|
NME/NM23 family member 5 |
| chr4_-_99435134 | 0.24 |
ENST00000476959.5
ENST00000482593.5 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr1_-_151175966 | 0.24 |
ENST00000441701.1
ENST00000295314.9 |
TMOD4
|
tropomodulin 4 |
| chr14_-_21098848 | 0.24 |
ENST00000556174.5
ENST00000554478.5 ENST00000553980.1 ENST00000421093.6 |
ZNF219
|
zinc finger protein 219 |
| chr7_+_148590760 | 0.23 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr3_-_98522869 | 0.22 |
ENST00000502288.5
ENST00000512147.5 ENST00000341181.11 ENST00000510541.5 ENST00000503621.5 ENST00000511081.5 ENST00000507874.5 ENST00000502299.5 ENST00000508659.5 ENST00000510545.5 ENST00000511667.5 ENST00000394185.6 |
CLDND1
|
claudin domain containing 1 |
| chr1_+_15341744 | 0.22 |
ENST00000444385.5
|
FHAD1
|
forkhead associated phosphopeptide binding domain 1 |
| chr16_+_53099100 | 0.21 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr6_-_69699124 | 0.21 |
ENST00000651675.1
|
LMBRD1
|
LMBR1 domain containing 1 |
| chr12_-_118190510 | 0.20 |
ENST00000540561.5
ENST00000537952.1 ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr6_+_72216442 | 0.20 |
ENST00000425662.6
ENST00000453976.6 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr7_+_90709530 | 0.20 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14 |
| chr17_-_73232218 | 0.20 |
ENST00000583024.1
ENST00000403627.7 ENST00000405159.7 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104 member A |
| chr5_-_135399863 | 0.20 |
ENST00000510038.1
ENST00000304332.8 |
MACROH2A1
|
macroH2A.1 histone |
| chr4_-_89029881 | 0.20 |
ENST00000506913.1
|
FAM13A
|
family with sequence similarity 13 member A |
| chr2_-_55010348 | 0.20 |
ENST00000394609.6
|
RTN4
|
reticulon 4 |
| chr4_-_142305826 | 0.19 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr4_+_68447453 | 0.19 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr16_+_81645352 | 0.19 |
ENST00000398040.8
|
CMIP
|
c-Maf inducing protein |
| chr14_-_80959484 | 0.19 |
ENST00000555529.5
ENST00000556042.5 ENST00000556981.5 |
CEP128
|
centrosomal protein 128 |
| chr17_+_70075215 | 0.19 |
ENST00000283936.5
ENST00000615244.4 ENST00000392671.6 |
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr12_+_75391078 | 0.19 |
ENST00000550916.6
ENST00000378692.7 ENST00000320460.8 ENST00000547164.1 |
GLIPR1L2
|
GLIPR1 like 2 |
| chr2_-_171231314 | 0.18 |
ENST00000521943.5
|
TLK1
|
tousled like kinase 1 |
| chr5_+_69565122 | 0.18 |
ENST00000507595.1
|
GTF2H2C
|
GTF2H2 family member C |
| chr3_-_151316795 | 0.18 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr3_-_12545499 | 0.18 |
ENST00000564146.4
|
MKRN2OS
|
MKRN2 opposite strand |
| chr6_+_27824084 | 0.18 |
ENST00000355057.3
|
H4C11
|
H4 clustered histone 11 |
| chr7_-_123199960 | 0.17 |
ENST00000194130.7
|
SLC13A1
|
solute carrier family 13 member 1 |
| chr1_+_74235377 | 0.17 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr7_+_142332182 | 0.17 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr17_+_70075317 | 0.16 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chr15_+_75336053 | 0.16 |
ENST00000564815.5
ENST00000338995.10 ENST00000267935.13 |
COMMD4
|
COMM domain containing 4 |
| chr20_+_31547367 | 0.15 |
ENST00000394552.3
|
MCTS2P
|
MCTS family member 2, pseudogene |
| chr11_-_124310837 | 0.15 |
ENST00000357821.2
|
OR8D1
|
olfactory receptor family 8 subfamily D member 1 |
| chr12_-_10172117 | 0.15 |
ENST00000545927.5
ENST00000309539.8 ENST00000432556.6 ENST00000544577.5 |
OLR1
|
oxidized low density lipoprotein receptor 1 |
| chr5_-_11589019 | 0.15 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2 |
| chr11_-_40294089 | 0.14 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr3_-_142029108 | 0.14 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr15_+_75336078 | 0.14 |
ENST00000567377.5
ENST00000562789.5 ENST00000568301.1 |
COMMD4
|
COMM domain containing 4 |
| chr9_+_65700287 | 0.13 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr5_-_9630351 | 0.13 |
ENST00000382492.4
|
TAS2R1
|
taste 2 receptor member 1 |
| chr12_-_11134644 | 0.13 |
ENST00000539585.1
|
TAS2R30
|
taste 2 receptor member 30 |
| chr3_+_98168700 | 0.13 |
ENST00000383696.4
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15 |
| chr12_+_8513499 | 0.13 |
ENST00000299665.3
|
CLEC4D
|
C-type lectin domain family 4 member D |
| chr2_+_44275473 | 0.13 |
ENST00000260649.11
|
SLC3A1
|
solute carrier family 3 member 1 |
| chr2_-_229921316 | 0.13 |
ENST00000428959.5
ENST00000675423.1 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr6_+_3258914 | 0.12 |
ENST00000438998.7
ENST00000419065.6 ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome assembly chaperone 4 |
| chrX_-_111410420 | 0.12 |
ENST00000371993.7
ENST00000680476.1 |
DCX
|
doublecortin |
| chr1_+_115029823 | 0.12 |
ENST00000256592.3
|
TSHB
|
thyroid stimulating hormone subunit beta |
| chr17_+_73232601 | 0.12 |
ENST00000359042.6
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr11_-_96343170 | 0.12 |
ENST00000524717.6
|
MAML2
|
mastermind like transcriptional coactivator 2 |
| chr1_-_186461089 | 0.12 |
ENST00000391997.3
|
PDC
|
phosducin |
| chr19_+_49513353 | 0.11 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr12_-_118359105 | 0.11 |
ENST00000541186.5
ENST00000539872.5 |
TAOK3
|
TAO kinase 3 |
| chr6_+_3258888 | 0.11 |
ENST00000380305.4
|
PSMG4
|
proteasome assembly chaperone 4 |
| chr9_+_27524285 | 0.11 |
ENST00000276943.3
|
IFNK
|
interferon kappa |
| chr1_-_219613069 | 0.10 |
ENST00000367211.6
ENST00000651890.1 |
ZC3H11B
|
zinc finger CCCH-type containing 11B |
| chr17_+_73232400 | 0.10 |
ENST00000535032.7
ENST00000577615.5 ENST00000585109.5 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr5_+_140827950 | 0.10 |
ENST00000378126.4
ENST00000529310.6 ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr7_+_90709231 | 0.10 |
ENST00000446790.5
ENST00000265741.7 |
CDK14
|
cyclin dependent kinase 14 |
| chrY_-_6874027 | 0.09 |
ENST00000215479.10
|
AMELY
|
amelogenin Y-linked |
| chr11_-_58731968 | 0.09 |
ENST00000278400.3
|
GLYAT
|
glycine-N-acyltransferase |
| chr2_-_208129824 | 0.09 |
ENST00000282141.4
|
CRYGC
|
crystallin gamma C |
| chr3_-_33645253 | 0.09 |
ENST00000333778.10
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr2_+_165294031 | 0.08 |
ENST00000283256.10
|
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr17_+_77185210 | 0.08 |
ENST00000431431.6
|
SEC14L1
|
SEC14 like lipid binding 1 |
| chr6_+_42929127 | 0.08 |
ENST00000394142.7
|
CNPY3
|
canopy FGF signaling regulator 3 |
| chr3_-_157160751 | 0.08 |
ENST00000461804.5
|
CCNL1
|
cyclin L1 |
| chr14_+_20110739 | 0.08 |
ENST00000641386.2
ENST00000641633.2 |
OR4K17
|
olfactory receptor family 4 subfamily K member 17 |
| chr1_-_241999091 | 0.08 |
ENST00000357246.4
|
MAP1LC3C
|
microtubule associated protein 1 light chain 3 gamma |
| chr1_-_209784521 | 0.08 |
ENST00000294811.2
|
C1orf74
|
chromosome 1 open reading frame 74 |
| chr7_+_90709816 | 0.07 |
ENST00000436577.3
|
CDK14
|
cyclin dependent kinase 14 |
| chr5_-_175961324 | 0.07 |
ENST00000432305.6
ENST00000505969.1 |
THOC3
|
THO complex 3 |
| chr3_-_98523013 | 0.06 |
ENST00000394181.6
ENST00000508902.5 ENST00000394180.6 |
CLDND1
|
claudin domain containing 1 |
| chr4_-_142305935 | 0.06 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chrX_+_120604199 | 0.06 |
ENST00000371315.3
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr4_+_48483324 | 0.06 |
ENST00000273861.5
|
SLC10A4
|
solute carrier family 10 member 4 |
| chr3_+_12351493 | 0.05 |
ENST00000683699.1
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr15_-_43106022 | 0.05 |
ENST00000627960.1
ENST00000290650.9 |
UBR1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chrX_+_108044967 | 0.05 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr6_-_32941018 | 0.05 |
ENST00000418107.3
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr17_-_13017952 | 0.05 |
ENST00000578071.1
ENST00000426905.7 ENST00000395962.6 ENST00000338034.9 ENST00000583371.5 |
ELAC2
|
elaC ribonuclease Z 2 |
| chr9_-_27005659 | 0.05 |
ENST00000380055.6
|
LRRC19
|
leucine rich repeat containing 19 |
| chr12_-_110445540 | 0.05 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex subunit 3 |
| chr17_+_55264952 | 0.05 |
ENST00000226067.10
|
HLF
|
HLF transcription factor, PAR bZIP family member |
| chr11_+_89924064 | 0.04 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr14_+_75069577 | 0.04 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger C2HC-type containing 1C |
| chr16_+_86566821 | 0.04 |
ENST00000649859.1
|
FOXC2
|
forkhead box C2 |
| chr5_-_77638713 | 0.04 |
ENST00000306422.5
|
OTP
|
orthopedia homeobox |
| chr2_+_75646775 | 0.04 |
ENST00000393909.7
ENST00000358788.10 ENST00000409374.5 |
MRPL19
|
mitochondrial ribosomal protein L19 |
| chrX_+_35919725 | 0.04 |
ENST00000297866.9
ENST00000378653.8 |
CFAP47
|
cilia and flagella associated protein 47 |
| chr9_+_21440437 | 0.04 |
ENST00000276927.3
|
IFNA1
|
interferon alpha 1 |
| chr3_+_42936859 | 0.04 |
ENST00000446977.2
ENST00000418176.1 |
ENSG00000273291.5
KRBOX1
|
novel protein KRAB box domain containing 1 |
| chr1_+_67166448 | 0.03 |
ENST00000347310.10
|
IL23R
|
interleukin 23 receptor |
| chr7_+_136868622 | 0.03 |
ENST00000680005.1
ENST00000445907.6 |
CHRM2
|
cholinergic receptor muscarinic 2 |
| chr14_+_22516273 | 0.03 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr3_+_2892199 | 0.03 |
ENST00000397459.6
|
CNTN4
|
contactin 4 |
| chr4_+_87832917 | 0.03 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.8 | 2.4 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.6 | 4.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.5 | 1.5 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.5 | 1.4 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.4 | 1.7 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.3 | 1.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 0.7 | GO:0071284 | copper ion export(GO:0060003) cellular response to lead ion(GO:0071284) |
| 0.2 | 0.7 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 4.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 0.8 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.8 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.3 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 1.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 1.0 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 1.0 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.3 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 0.3 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 2.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.3 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 1.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 1.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.2 | GO:0038016 | insulin receptor internalization(GO:0038016) |
| 0.0 | 0.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 1.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.3 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.7 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 1.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.3 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 1.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.0 | GO:1990180 | tRNA 3'-trailer cleavage, endonucleolytic(GO:0034414) tRNA 3'-trailer cleavage(GO:0042779) mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.8 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 4.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 1.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 1.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.8 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.3 | 1.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 1.2 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.3 | 4.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 0.7 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 0.7 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 1.7 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.2 | 1.8 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 1.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 0.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 2.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.3 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.1 | 0.3 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.1 | 0.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 1.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.3 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 1.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 1.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.8 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) co-receptor binding(GO:0039706) |
| 0.0 | 1.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.0 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 1.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 4.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 3.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.1 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.2 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |