Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
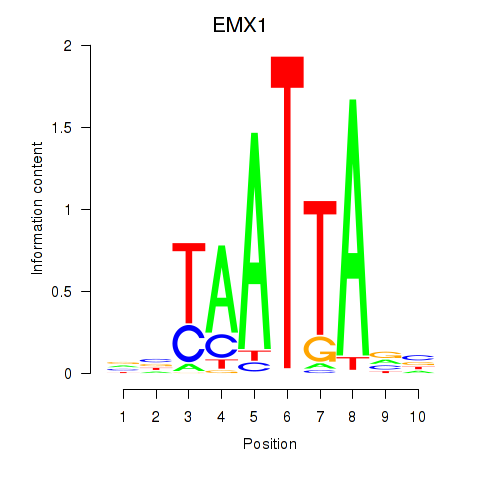
Results for EMX1
Z-value: 0.52
Transcription factors associated with EMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX1
|
ENSG00000135638.14 | EMX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX1 | hg38_v1_chr2_+_72917489_72917525, hg38_v1_chr2_+_72916183_72916260 | 0.31 | 1.3e-01 | Click! |
Activity profile of EMX1 motif
Sorted Z-values of EMX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_76023489 | 0.61 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10 |
| chr4_+_94995919 | 0.61 |
ENST00000509540.6
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr6_+_29111560 | 0.54 |
ENST00000377169.2
|
OR2J3
|
olfactory receptor family 2 subfamily J member 3 |
| chr21_-_30881572 | 0.51 |
ENST00000332378.6
|
KRTAP11-1
|
keratin associated protein 11-1 |
| chr3_+_98149326 | 0.50 |
ENST00000437310.1
|
OR5H14
|
olfactory receptor family 5 subfamily H member 14 |
| chr10_-_49762335 | 0.48 |
ENST00000419399.4
ENST00000432695.2 |
OGDHL
|
oxoglutarate dehydrogenase L |
| chr6_-_169250825 | 0.47 |
ENST00000676869.1
ENST00000676760.1 |
THBS2
|
thrombospondin 2 |
| chr3_+_42979281 | 0.47 |
ENST00000488863.5
ENST00000430121.3 |
GASK1A
|
golgi associated kinase 1A |
| chr3_-_142029108 | 0.47 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr10_-_49762276 | 0.46 |
ENST00000374103.9
|
OGDHL
|
oxoglutarate dehydrogenase L |
| chr15_+_76336755 | 0.45 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr20_-_51802433 | 0.43 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr12_+_26195647 | 0.41 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr7_-_93148345 | 0.40 |
ENST00000437805.5
ENST00000446959.5 ENST00000439952.5 ENST00000414791.5 ENST00000446033.1 ENST00000411955.5 ENST00000318238.9 |
SAMD9L
|
sterile alpha motif domain containing 9 like |
| chr5_-_151093566 | 0.39 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr4_+_85604146 | 0.38 |
ENST00000512201.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr8_+_49911396 | 0.37 |
ENST00000642720.2
|
SNTG1
|
syntrophin gamma 1 |
| chr4_+_94996232 | 0.36 |
ENST00000512312.5
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr7_-_73624492 | 0.36 |
ENST00000414749.6
ENST00000429400.6 ENST00000434326.5 ENST00000313375.8 ENST00000354613.5 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein like |
| chr8_+_49911604 | 0.36 |
ENST00000642164.1
ENST00000644093.1 ENST00000643999.1 ENST00000647073.1 ENST00000646880.1 |
SNTG1
|
syntrophin gamma 1 |
| chr6_-_52840843 | 0.36 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr10_-_114144599 | 0.36 |
ENST00000428953.1
|
CCDC186
|
coiled-coil domain containing 186 |
| chr9_-_16728165 | 0.36 |
ENST00000603713.5
ENST00000603313.5 |
BNC2
|
basonuclin 2 |
| chr11_-_117877463 | 0.36 |
ENST00000527717.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr12_+_80716906 | 0.35 |
ENST00000228644.4
|
MYF5
|
myogenic factor 5 |
| chr6_-_49713564 | 0.35 |
ENST00000616725.4
ENST00000618917.4 |
CRISP2
|
cysteine rich secretory protein 2 |
| chr6_-_49713521 | 0.35 |
ENST00000339139.5
|
CRISP2
|
cysteine rich secretory protein 2 |
| chr11_-_117876719 | 0.34 |
ENST00000529335.6
ENST00000260282.8 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr17_-_41149823 | 0.33 |
ENST00000343246.6
|
KRTAP4-5
|
keratin associated protein 4-5 |
| chr8_+_22567038 | 0.33 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr11_-_117876892 | 0.33 |
ENST00000539526.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr8_-_42377227 | 0.33 |
ENST00000220812.3
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr20_+_61599755 | 0.32 |
ENST00000543233.2
|
CDH4
|
cadherin 4 |
| chr11_+_92969651 | 0.31 |
ENST00000257068.3
ENST00000528076.1 |
MTNR1B
|
melatonin receptor 1B |
| chr5_+_132873660 | 0.31 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chr16_-_81220370 | 0.30 |
ENST00000337114.8
|
PKD1L2
|
polycystin 1 like 2 (gene/pseudogene) |
| chr19_+_15728024 | 0.29 |
ENST00000305899.5
|
OR10H2
|
olfactory receptor family 10 subfamily H member 2 |
| chr17_-_41034871 | 0.29 |
ENST00000344363.7
|
KRTAP1-3
|
keratin associated protein 1-3 |
| chr12_-_91153149 | 0.28 |
ENST00000550758.1
|
DCN
|
decorin |
| chr11_-_101129706 | 0.28 |
ENST00000534013.5
|
PGR
|
progesterone receptor |
| chr1_+_175067831 | 0.28 |
ENST00000239462.9
|
TNN
|
tenascin N |
| chr19_-_3557563 | 0.28 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr1_+_152908538 | 0.27 |
ENST00000368764.4
|
IVL
|
involucrin |
| chr8_-_56113982 | 0.27 |
ENST00000311923.1
|
MOS
|
MOS proto-oncogene, serine/threonine kinase |
| chr21_+_30396030 | 0.27 |
ENST00000355459.4
|
KRTAP13-1
|
keratin associated protein 13-1 |
| chr15_+_62561361 | 0.27 |
ENST00000561311.5
|
TLN2
|
talin 2 |
| chr18_+_61333424 | 0.27 |
ENST00000262717.9
|
CDH20
|
cadherin 20 |
| chr10_-_73591330 | 0.26 |
ENST00000451492.5
ENST00000681793.1 ENST00000680396.1 ENST00000413442.5 |
USP54
|
ubiquitin specific peptidase 54 |
| chr6_-_25042003 | 0.26 |
ENST00000510784.8
|
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr2_+_168901290 | 0.26 |
ENST00000429379.2
ENST00000375363.8 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase catalytic subunit 2 |
| chr4_-_73620391 | 0.24 |
ENST00000395777.6
ENST00000307439.10 |
RASSF6
|
Ras association domain family member 6 |
| chr11_-_35360050 | 0.24 |
ENST00000644868.1
ENST00000643454.1 ENST00000646080.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr1_-_150765735 | 0.24 |
ENST00000679898.1
ENST00000448301.7 ENST00000680664.1 ENST00000679512.1 ENST00000368985.8 ENST00000679582.1 |
CTSS
|
cathepsin S |
| chr19_+_12938598 | 0.23 |
ENST00000586760.2
ENST00000316448.10 ENST00000588454.6 |
CALR
|
calreticulin |
| chr7_-_81770122 | 0.23 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor |
| chr6_-_22297028 | 0.23 |
ENST00000306482.2
|
PRL
|
prolactin |
| chr4_+_70334963 | 0.22 |
ENST00000273936.6
|
CABS1
|
calcium binding protein, spermatid associated 1 |
| chr5_+_172641241 | 0.22 |
ENST00000369800.6
ENST00000520919.5 ENST00000522853.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr14_+_21990357 | 0.22 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr14_+_67819761 | 0.22 |
ENST00000487861.5
|
RAD51B
|
RAD51 paralog B |
| chr1_-_152414256 | 0.22 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr11_+_5383812 | 0.21 |
ENST00000642046.1
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr19_-_45584810 | 0.21 |
ENST00000323060.3
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3 |
| chr8_+_49911801 | 0.21 |
ENST00000643809.1
|
SNTG1
|
syntrophin gamma 1 |
| chr12_+_18261511 | 0.20 |
ENST00000538779.6
ENST00000433979.6 |
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr7_+_142399860 | 0.20 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1 |
| chr2_-_208146150 | 0.20 |
ENST00000260988.5
|
CRYGB
|
crystallin gamma B |
| chr1_-_92486916 | 0.20 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr5_+_141387698 | 0.20 |
ENST00000615384.1
ENST00000519479.2 |
PCDHGB4
|
protocadherin gamma subfamily B, 4 |
| chr18_-_36129305 | 0.20 |
ENST00000269187.10
ENST00000590986.5 ENST00000440549.6 |
SLC39A6
|
solute carrier family 39 member 6 |
| chr7_+_44606563 | 0.20 |
ENST00000439616.6
|
OGDH
|
oxoglutarate dehydrogenase |
| chr18_-_36122110 | 0.19 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 member 6 |
| chr3_-_33645433 | 0.19 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr8_-_85341705 | 0.19 |
ENST00000517618.5
|
CA1
|
carbonic anhydrase 1 |
| chr19_-_14835252 | 0.19 |
ENST00000641666.1
ENST00000642030.1 ENST00000642000.1 |
OR7C1
|
olfactory receptor family 7 subfamily C member 1 |
| chr16_+_57976435 | 0.19 |
ENST00000290871.10
ENST00000441824.4 |
TEPP
|
testis, prostate and placenta expressed |
| chr3_+_114294020 | 0.19 |
ENST00000383671.8
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains |
| chr4_+_94974984 | 0.19 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr1_+_160190567 | 0.19 |
ENST00000368078.8
|
CASQ1
|
calsequestrin 1 |
| chr19_+_15082211 | 0.18 |
ENST00000641398.1
|
OR1I1
|
olfactory receptor family 1 subfamily I member 1 |
| chr19_+_37371152 | 0.18 |
ENST00000483919.5
ENST00000588911.5 ENST00000587349.1 |
ZNF527
|
zinc finger protein 527 |
| chr5_-_143400716 | 0.18 |
ENST00000424646.6
ENST00000652686.1 |
NR3C1
|
nuclear receptor subfamily 3 group C member 1 |
| chr21_-_42395943 | 0.18 |
ENST00000398405.5
|
TMPRSS3
|
transmembrane serine protease 3 |
| chr7_-_88795732 | 0.18 |
ENST00000297203.3
|
TEX47
|
testis expressed 47 |
| chr11_-_101129806 | 0.18 |
ENST00000325455.10
ENST00000617858.4 ENST00000619228.2 |
PGR
|
progesterone receptor |
| chr3_-_151316795 | 0.18 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr16_-_29923237 | 0.18 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr10_+_24208774 | 0.18 |
ENST00000376456.8
ENST00000458595.5 ENST00000376452.7 ENST00000430453.6 |
KIAA1217
|
KIAA1217 |
| chr1_-_150765785 | 0.18 |
ENST00000680311.1
ENST00000681728.1 ENST00000680288.1 |
CTSS
|
cathepsin S |
| chr17_-_48722510 | 0.18 |
ENST00000290294.5
|
PRAC1
|
PRAC1 small nuclear protein |
| chr2_+_124025280 | 0.18 |
ENST00000431078.1
ENST00000682447.1 |
CNTNAP5
|
contactin associated protein family member 5 |
| chr11_-_36598221 | 0.17 |
ENST00000311485.8
ENST00000527033.5 ENST00000532616.1 ENST00000618712.4 |
RAG2
|
recombination activating 2 |
| chr18_+_616672 | 0.17 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1 |
| chr1_+_159204860 | 0.17 |
ENST00000368122.4
ENST00000368121.6 |
ACKR1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr16_+_7303245 | 0.17 |
ENST00000674626.1
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr6_-_32941018 | 0.17 |
ENST00000418107.3
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr8_+_109086585 | 0.17 |
ENST00000518632.2
|
TRHR
|
thyrotropin releasing hormone receptor |
| chr6_+_131250375 | 0.17 |
ENST00000474850.2
|
AKAP7
|
A-kinase anchoring protein 7 |
| chrX_+_37990773 | 0.17 |
ENST00000341016.5
|
H2AP
|
H2A.P histone |
| chr2_-_165203870 | 0.17 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr14_+_21997531 | 0.17 |
ENST00000390445.2
|
TRAV17
|
T cell receptor alpha variable 17 |
| chr5_+_90640718 | 0.16 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr6_+_29306626 | 0.16 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr6_-_46325641 | 0.16 |
ENST00000330430.10
ENST00000405162.2 |
RCAN2
|
regulator of calcineurin 2 |
| chr17_+_81683963 | 0.16 |
ENST00000676462.1
ENST00000679336.1 ENST00000678196.1 ENST00000677243.1 ENST00000677044.1 ENST00000677109.1 ENST00000677484.1 ENST00000678105.1 ENST00000677209.1 ENST00000329138.9 ENST00000677225.1 ENST00000678866.1 ENST00000676729.1 ENST00000572392.2 ENST00000577012.1 |
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr6_-_33289189 | 0.16 |
ENST00000374617.9
|
WDR46
|
WD repeat domain 46 |
| chrX_-_70260199 | 0.16 |
ENST00000374519.4
|
P2RY4
|
pyrimidinergic receptor P2Y4 |
| chr7_+_148590760 | 0.16 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr5_+_98773651 | 0.16 |
ENST00000513185.3
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr10_+_17951825 | 0.16 |
ENST00000539911.5
|
SLC39A12
|
solute carrier family 39 member 12 |
| chr10_+_17951885 | 0.15 |
ENST00000377374.8
|
SLC39A12
|
solute carrier family 39 member 12 |
| chr7_-_101217569 | 0.15 |
ENST00000223127.8
|
PLOD3
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 |
| chr8_-_40897814 | 0.15 |
ENST00000297737.11
ENST00000315769.11 |
ZMAT4
|
zinc finger matrin-type 4 |
| chr10_+_17951906 | 0.15 |
ENST00000377371.3
ENST00000377369.7 |
SLC39A12
|
solute carrier family 39 member 12 |
| chrX_+_38352573 | 0.15 |
ENST00000039007.5
|
OTC
|
ornithine transcarbamylase |
| chr6_+_63521738 | 0.15 |
ENST00000648894.1
ENST00000639568.2 |
PTP4A1
|
protein tyrosine phosphatase 4A1 |
| chr14_+_32329341 | 0.15 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr3_-_131026726 | 0.15 |
ENST00000514044.5
ENST00000264992.8 |
ASTE1
|
asteroid homolog 1 |
| chr6_+_127577168 | 0.15 |
ENST00000329722.8
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr7_+_70596078 | 0.15 |
ENST00000644506.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2 |
| chr4_+_168497066 | 0.15 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_-_23584600 | 0.15 |
ENST00000396007.6
|
SOX5
|
SRY-box transcription factor 5 |
| chr7_-_122699108 | 0.14 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133 |
| chr6_+_26365215 | 0.14 |
ENST00000527422.5
ENST00000356386.6 ENST00000396948.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr11_+_5389377 | 0.14 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr15_+_58987652 | 0.14 |
ENST00000348370.9
ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr4_+_2418932 | 0.14 |
ENST00000635017.1
|
CFAP99
|
cilia and flagella associated protein 99 |
| chr12_-_101210232 | 0.14 |
ENST00000536262.3
|
SLC5A8
|
solute carrier family 5 member 8 |
| chr19_-_51019699 | 0.14 |
ENST00000358789.8
|
KLK10
|
kallikrein related peptidase 10 |
| chr12_+_6904733 | 0.14 |
ENST00000007969.12
ENST00000622489.4 ENST00000443597.7 ENST00000323702.9 |
LRRC23
|
leucine rich repeat containing 23 |
| chr17_-_40994159 | 0.14 |
ENST00000391586.3
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr11_-_117876612 | 0.14 |
ENST00000584230.1
ENST00000526014.6 ENST00000584394.5 ENST00000614497.5 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr4_-_142305826 | 0.14 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr1_+_154429315 | 0.14 |
ENST00000476006.5
|
IL6R
|
interleukin 6 receptor |
| chr2_-_74392025 | 0.14 |
ENST00000440727.1
ENST00000409240.5 |
DCTN1
|
dynactin subunit 1 |
| chr20_-_51802509 | 0.14 |
ENST00000371539.7
ENST00000217086.9 |
SALL4
|
spalt like transcription factor 4 |
| chr12_-_52618559 | 0.14 |
ENST00000305748.7
|
KRT73
|
keratin 73 |
| chr5_+_31193739 | 0.14 |
ENST00000514738.5
|
CDH6
|
cadherin 6 |
| chr19_+_18831951 | 0.14 |
ENST00000262803.10
ENST00000599848.5 |
UPF1
|
UPF1 RNA helicase and ATPase |
| chr17_+_4807119 | 0.13 |
ENST00000263088.11
ENST00000572940.5 |
PLD2
|
phospholipase D2 |
| chr6_+_26402237 | 0.13 |
ENST00000476549.6
ENST00000450085.6 ENST00000425234.6 ENST00000427334.5 ENST00000506698.1 ENST00000289361.11 |
BTN3A1
|
butyrophilin subfamily 3 member A1 |
| chr14_+_100019375 | 0.13 |
ENST00000544450.6
|
EVL
|
Enah/Vasp-like |
| chr7_-_100119840 | 0.13 |
ENST00000437822.6
|
TAF6
|
TATA-box binding protein associated factor 6 |
| chr5_-_20575850 | 0.13 |
ENST00000507958.5
|
CDH18
|
cadherin 18 |
| chr4_-_68245683 | 0.13 |
ENST00000332644.6
|
TMPRSS11B
|
transmembrane serine protease 11B |
| chr4_-_115113614 | 0.13 |
ENST00000264363.7
|
NDST4
|
N-deacetylase and N-sulfotransferase 4 |
| chr12_+_26195313 | 0.13 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr7_-_44189428 | 0.13 |
ENST00000673284.1
ENST00000403799.8 ENST00000671824.1 |
GCK
|
glucokinase |
| chr2_+_90209873 | 0.13 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr8_+_32721823 | 0.13 |
ENST00000539990.3
ENST00000519240.5 |
NRG1
|
neuregulin 1 |
| chr2_+_90100235 | 0.13 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chrX_+_30235894 | 0.13 |
ENST00000620842.1
|
MAGEB3
|
MAGE family member B3 |
| chr5_-_24644968 | 0.13 |
ENST00000264463.8
|
CDH10
|
cadherin 10 |
| chr4_+_168497044 | 0.13 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_-_89100352 | 0.13 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr4_-_115113822 | 0.13 |
ENST00000613194.4
|
NDST4
|
N-deacetylase and N-sulfotransferase 4 |
| chr19_-_14979848 | 0.13 |
ENST00000594383.2
|
SLC1A6
|
solute carrier family 1 member 6 |
| chr11_+_67452392 | 0.13 |
ENST00000438189.6
|
CABP4
|
calcium binding protein 4 |
| chr15_-_55270280 | 0.12 |
ENST00000564609.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr18_+_23992773 | 0.12 |
ENST00000304621.10
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr21_+_42403874 | 0.12 |
ENST00000319294.11
ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chrX_-_15314543 | 0.12 |
ENST00000344384.8
|
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr22_-_30326923 | 0.12 |
ENST00000215790.12
|
TBC1D10A
|
TBC1 domain family member 10A |
| chr12_-_10826358 | 0.12 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chr4_+_99511008 | 0.12 |
ENST00000514652.5
ENST00000326581.9 |
C4orf17
|
chromosome 4 open reading frame 17 |
| chr9_-_21207143 | 0.12 |
ENST00000357374.2
|
IFNA10
|
interferon alpha 10 |
| chr8_+_38974212 | 0.12 |
ENST00000302495.5
|
HTRA4
|
HtrA serine peptidase 4 |
| chrM_+_12329 | 0.12 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 5 |
| chr16_-_29899245 | 0.12 |
ENST00000537485.5
|
SEZ6L2
|
seizure related 6 homolog like 2 |
| chr1_-_88891496 | 0.12 |
ENST00000448623.5
ENST00000370500.10 ENST00000418217.1 |
GTF2B
|
general transcription factor IIB |
| chr13_+_45464901 | 0.12 |
ENST00000349995.10
|
COG3
|
component of oligomeric golgi complex 3 |
| chr11_-_117929380 | 0.12 |
ENST00000528626.5
ENST00000445164.6 ENST00000430170.6 ENST00000524993.6 ENST00000526090.1 |
TMPRSS13
|
transmembrane serine protease 13 |
| chr16_+_24729692 | 0.12 |
ENST00000315183.11
|
TNRC6A
|
trinucleotide repeat containing adaptor 6A |
| chr13_-_37598750 | 0.12 |
ENST00000379743.8
ENST00000379742.4 ENST00000379749.8 ENST00000379747.9 ENST00000541179.5 ENST00000541481.5 |
POSTN
|
periostin |
| chr8_+_104339796 | 0.12 |
ENST00000622554.1
ENST00000297581.2 |
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr11_-_96343170 | 0.12 |
ENST00000524717.6
|
MAML2
|
mastermind like transcriptional coactivator 2 |
| chr20_+_33079640 | 0.12 |
ENST00000375483.4
|
BPIFB4
|
BPI fold containing family B member 4 |
| chr5_-_142698004 | 0.12 |
ENST00000407758.5
ENST00000441680.6 ENST00000419524.6 ENST00000621536.4 |
FGF1
|
fibroblast growth factor 1 |
| chr12_+_130953898 | 0.12 |
ENST00000261654.10
|
ADGRD1
|
adhesion G protein-coupled receptor D1 |
| chr4_+_105710809 | 0.11 |
ENST00000360505.9
ENST00000510865.5 ENST00000509336.5 |
GSTCD
|
glutathione S-transferase C-terminal domain containing |
| chr2_-_74507664 | 0.11 |
ENST00000233630.11
|
PCGF1
|
polycomb group ring finger 1 |
| chr20_-_290717 | 0.11 |
ENST00000360321.7
ENST00000400269.4 |
C20orf96
|
chromosome 20 open reading frame 96 |
| chr22_-_32255344 | 0.11 |
ENST00000266086.6
|
SLC5A4
|
solute carrier family 5 member 4 |
| chr3_+_178419123 | 0.11 |
ENST00000614557.1
ENST00000455307.5 ENST00000436432.1 |
ENSG00000275163.1
LINC01014
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 (KCNMB2-IT1 - KCNMB2 readthrough transcript) long intergenic non-protein coding RNA 1014 |
| chr6_+_26365176 | 0.11 |
ENST00000377708.7
|
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr2_+_27275429 | 0.11 |
ENST00000420191.5
ENST00000296097.8 |
DNAJC5G
|
DnaJ heat shock protein family (Hsp40) member C5 gamma |
| chr13_-_36214584 | 0.11 |
ENST00000317764.6
|
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr11_-_105035113 | 0.11 |
ENST00000526568.5
ENST00000531166.5 ENST00000534497.5 ENST00000527979.5 ENST00000533400.6 ENST00000528974.1 ENST00000525825.5 ENST00000353247.9 ENST00000446369.5 ENST00000436863.7 |
CASP1
|
caspase 1 |
| chr10_-_28282086 | 0.11 |
ENST00000375719.7
ENST00000375732.5 |
MPP7
|
membrane palmitoylated protein 7 |
| chr3_+_148730100 | 0.11 |
ENST00000474935.5
ENST00000475347.5 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor type 1 |
| chr17_+_32021005 | 0.11 |
ENST00000327564.11
ENST00000584368.5 ENST00000394713.7 ENST00000341671.11 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr11_+_5449323 | 0.11 |
ENST00000641930.1
|
OR51I2
|
olfactory receptor family 51 subfamily I member 2 |
| chr6_+_42155399 | 0.11 |
ENST00000623004.2
ENST00000372963.4 ENST00000654459.1 |
GUCA1ANB
GUCA1A
|
GUCA1A neighbor guanylate cyclase activator 1A |
| chr2_-_24328113 | 0.11 |
ENST00000622089.4
|
ITSN2
|
intersectin 2 |
| chr6_-_31139063 | 0.11 |
ENST00000259845.5
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr18_+_24460630 | 0.11 |
ENST00000256906.5
|
HRH4
|
histamine receptor H4 |
| chr14_+_69879408 | 0.11 |
ENST00000361956.8
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr1_-_44843240 | 0.11 |
ENST00000372192.4
|
PTCH2
|
patched 2 |
| chr11_-_57381146 | 0.11 |
ENST00000287143.2
|
PRG3
|
proteoglycan 3, pro eosinophil major basic protein 2 |
| chr3_-_193378747 | 0.10 |
ENST00000342358.9
|
ATP13A5
|
ATPase 13A5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.2 | 0.5 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.4 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.4 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.5 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.1 | 0.3 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.5 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 | 0.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.2 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.1 | 0.3 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 0.6 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.3 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.1 | 0.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.2 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0032761 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:2000798 | amniotic stem cell differentiation(GO:0097086) negative regulation of dense core granule biogenesis(GO:2000706) negative regulation of mesenchymal stem cell differentiation(GO:2000740) regulation of amniotic stem cell differentiation(GO:2000797) negative regulation of amniotic stem cell differentiation(GO:2000798) |
| 0.0 | 0.4 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 1.1 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:1900222 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0089712 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.3 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.2 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.5 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.0 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 1.2 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.4 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.9 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.4 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 0.2 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.2 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 1.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.3 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0052854 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 1.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.0 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.0 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |