Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
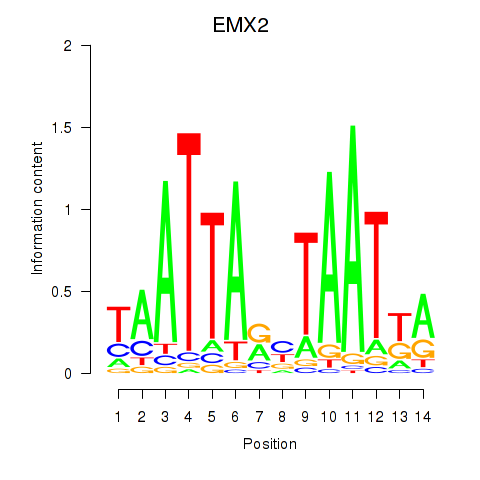
Results for EMX2
Z-value: 0.65
Transcription factors associated with EMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX2
|
ENSG00000170370.12 | EMX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX2 | hg38_v1_chr10_+_117543567_117543658, hg38_v1_chr10_+_117542416_117542445, hg38_v1_chr10_+_117542721_117542754 | 0.35 | 8.2e-02 | Click! |
Activity profile of EMX2 motif
Sorted Z-values of EMX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_84181630 | 3.02 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr4_+_41538143 | 2.65 |
ENST00000503057.6
ENST00000511496.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_84144260 | 2.07 |
ENST00000370685.7
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr4_-_100517991 | 1.95 |
ENST00000511970.5
ENST00000502569.1 ENST00000305864.7 ENST00000296420.9 |
EMCN
|
endomucin |
| chr7_+_80133830 | 1.95 |
ENST00000648098.1
ENST00000648476.1 ENST00000648412.1 ENST00000648953.1 ENST00000648306.1 ENST00000648832.1 ENST00000648877.1 ENST00000442586.2 ENST00000649487.1 ENST00000649267.1 |
GNAI1
|
G protein subunit alpha i1 |
| chr19_+_14583076 | 1.90 |
ENST00000547437.5
ENST00000417570.6 |
CLEC17A
|
C-type lectin domain containing 17A |
| chr2_-_98663464 | 1.87 |
ENST00000414521.6
|
MGAT4A
|
alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase A |
| chr6_-_75363003 | 1.64 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr4_-_185956652 | 1.45 |
ENST00000355634.9
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr10_-_91633057 | 1.33 |
ENST00000238994.6
|
PPP1R3C
|
protein phosphatase 1 regulatory subunit 3C |
| chr4_-_88284747 | 1.29 |
ENST00000514204.1
|
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent 1K |
| chr9_-_41908681 | 1.27 |
ENST00000476961.5
|
CNTNAP3B
|
contactin associated protein family member 3B |
| chr4_-_185956348 | 1.27 |
ENST00000431902.5
ENST00000284776.11 ENST00000415274.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_78004930 | 1.26 |
ENST00000370763.6
|
DNAJB4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr11_-_63608542 | 1.25 |
ENST00000540943.1
|
PLAAT3
|
phospholipase A and acyltransferase 3 |
| chr17_-_69141878 | 1.18 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr9_-_14314132 | 1.15 |
ENST00000380953.6
|
NFIB
|
nuclear factor I B |
| chr18_-_55423757 | 1.03 |
ENST00000675707.1
|
TCF4
|
transcription factor 4 |
| chr9_+_128566741 | 1.03 |
ENST00000630866.1
|
SPTAN1
|
spectrin alpha, non-erythrocytic 1 |
| chr14_+_24120956 | 0.95 |
ENST00000558325.2
|
ENSG00000259371.2
|
novel protein |
| chr12_-_24902243 | 0.92 |
ENST00000538118.5
|
BCAT1
|
branched chain amino acid transaminase 1 |
| chr5_-_111976925 | 0.90 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr1_+_77918128 | 0.89 |
ENST00000342754.5
|
NEXN
|
nexilin F-actin binding protein |
| chr6_-_110179995 | 0.87 |
ENST00000392586.5
ENST00000419252.1 ENST00000359451.6 ENST00000392588.5 |
WASF1
|
WASP family member 1 |
| chr6_+_135851681 | 0.87 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B |
| chr5_-_56116946 | 0.86 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr8_+_97887903 | 0.83 |
ENST00000520016.5
|
MATN2
|
matrilin 2 |
| chr4_-_137532452 | 0.77 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr11_-_125111708 | 0.76 |
ENST00000531909.5
ENST00000529530.1 |
TMEM218
|
transmembrane protein 218 |
| chr6_+_28124596 | 0.76 |
ENST00000340487.5
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr3_-_122793772 | 0.74 |
ENST00000306103.3
|
HSPBAP1
|
HSPB1 associated protein 1 |
| chr9_+_72577939 | 0.73 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr3_-_142029108 | 0.72 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr6_-_53510445 | 0.70 |
ENST00000509541.5
|
GCLC
|
glutamate-cysteine ligase catalytic subunit |
| chr1_-_160954801 | 0.69 |
ENST00000368029.4
|
ITLN2
|
intelectin 2 |
| chr12_+_59689337 | 0.69 |
ENST00000261187.8
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr9_-_21202205 | 0.66 |
ENST00000239347.3
|
IFNA7
|
interferon alpha 7 |
| chr5_-_140633690 | 0.66 |
ENST00000512545.1
ENST00000401743.6 |
CD14
|
CD14 molecule |
| chr12_-_57826295 | 0.66 |
ENST00000549039.5
|
CTDSP2
|
CTD small phosphatase 2 |
| chr5_-_9630351 | 0.66 |
ENST00000382492.4
|
TAS2R1
|
taste 2 receptor member 1 |
| chr3_-_142000353 | 0.64 |
ENST00000499676.5
|
TFDP2
|
transcription factor Dp-2 |
| chr17_-_445939 | 0.64 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr1_-_44986568 | 0.63 |
ENST00000372183.7
ENST00000360403.7 ENST00000620860.4 |
EIF2B3
|
eukaryotic translation initiation factor 2B subunit gamma |
| chr3_-_58537283 | 0.62 |
ENST00000459701.6
|
ACOX2
|
acyl-CoA oxidase 2 |
| chr1_+_28438104 | 0.61 |
ENST00000633167.1
ENST00000373836.4 |
PHACTR4
|
phosphatase and actin regulator 4 |
| chr3_-_58537181 | 0.61 |
ENST00000302819.10
|
ACOX2
|
acyl-CoA oxidase 2 |
| chr12_-_122716790 | 0.61 |
ENST00000528880.3
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr3_-_185821092 | 0.60 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr5_+_90640718 | 0.60 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr6_+_29111560 | 0.60 |
ENST00000377169.2
|
OR2J3
|
olfactory receptor family 2 subfamily J member 3 |
| chr3_+_132597260 | 0.59 |
ENST00000249887.3
|
ACKR4
|
atypical chemokine receptor 4 |
| chr12_-_68302872 | 0.59 |
ENST00000539972.5
|
MDM1
|
Mdm1 nuclear protein |
| chr3_+_113897470 | 0.57 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr17_+_68525795 | 0.57 |
ENST00000592800.5
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr4_-_25863537 | 0.57 |
ENST00000502949.5
ENST00000264868.9 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
SEL1L family member 3 |
| chr21_-_30497160 | 0.55 |
ENST00000334058.3
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr9_-_76906041 | 0.55 |
ENST00000443509.6
ENST00000428286.5 ENST00000376713.3 |
PRUNE2
|
prune homolog 2 with BCH domain |
| chr6_+_72216442 | 0.55 |
ENST00000425662.6
ENST00000453976.6 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr9_-_5833014 | 0.55 |
ENST00000339450.10
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr19_+_49513353 | 0.53 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chr17_-_55511434 | 0.52 |
ENST00000636752.1
|
SMIM36
|
small integral membrane protein 36 |
| chr7_+_116222804 | 0.48 |
ENST00000393481.6
|
TES
|
testin LIM domain protein |
| chr12_+_125065427 | 0.47 |
ENST00000316519.11
|
AACS
|
acetoacetyl-CoA synthetase |
| chr18_+_32190015 | 0.46 |
ENST00000581447.1
|
MEP1B
|
meprin A subunit beta |
| chr4_-_109801978 | 0.46 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr19_-_51417700 | 0.45 |
ENST00000529627.1
ENST00000439889.6 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr1_+_117001744 | 0.45 |
ENST00000256652.8
ENST00000682167.1 ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr20_-_31390580 | 0.44 |
ENST00000339144.3
ENST00000376321.4 |
DEFB119
|
defensin beta 119 |
| chr18_+_32190033 | 0.44 |
ENST00000269202.11
|
MEP1B
|
meprin A subunit beta |
| chr5_+_140875299 | 0.44 |
ENST00000613593.1
ENST00000398631.3 |
PCDHA12
|
protocadherin alpha 12 |
| chr1_+_162790702 | 0.44 |
ENST00000254521.8
ENST00000367915.1 |
HSD17B7
|
hydroxysteroid 17-beta dehydrogenase 7 |
| chrX_+_77910656 | 0.44 |
ENST00000343533.9
ENST00000341514.11 ENST00000645454.1 ENST00000642651.1 ENST00000644362.1 |
ATP7A
PGK1
|
ATPase copper transporting alpha phosphoglycerate kinase 1 |
| chr8_+_69492793 | 0.43 |
ENST00000616868.1
ENST00000419716.7 ENST00000402687.9 |
SULF1
|
sulfatase 1 |
| chr11_-_119196769 | 0.43 |
ENST00000415318.2
|
CCDC153
|
coiled-coil domain containing 153 |
| chr19_-_54941610 | 0.42 |
ENST00000328092.9
ENST00000590030.5 |
NLRP7
|
NLR family pyrin domain containing 7 |
| chr3_+_111674654 | 0.42 |
ENST00000636933.1
ENST00000393934.7 ENST00000477665.2 |
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2 |
| chr19_-_4558417 | 0.41 |
ENST00000586965.1
|
SEMA6B
|
semaphorin 6B |
| chr16_+_56961942 | 0.41 |
ENST00000200676.8
ENST00000566128.1 |
CETP
|
cholesteryl ester transfer protein |
| chr12_+_29149238 | 0.40 |
ENST00000536681.8
|
FAR2
|
fatty acyl-CoA reductase 2 |
| chr22_-_32464440 | 0.39 |
ENST00000397450.2
ENST00000397452.5 ENST00000300399.8 |
BPIFC
|
BPI fold containing family C |
| chr1_+_92168915 | 0.39 |
ENST00000637221.2
|
BTBD8
|
BTB domain containing 8 |
| chr11_+_74592567 | 0.38 |
ENST00000263681.7
ENST00000527458.5 ENST00000532497.5 ENST00000530511.5 |
POLD3
|
DNA polymerase delta 3, accessory subunit |
| chr11_+_119149029 | 0.37 |
ENST00000619701.5
|
ABCG4
|
ATP binding cassette subfamily G member 4 |
| chr11_-_60183191 | 0.37 |
ENST00000412309.6
|
MS4A6A
|
membrane spanning 4-domains A6A |
| chr1_+_15341744 | 0.36 |
ENST00000444385.5
|
FHAD1
|
forkhead associated phosphopeptide binding domain 1 |
| chr16_+_18983927 | 0.36 |
ENST00000569532.5
ENST00000304381.10 |
TMC7
|
transmembrane channel like 7 |
| chr6_+_72212802 | 0.35 |
ENST00000401910.7
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr7_+_107583919 | 0.35 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr3_+_159839847 | 0.35 |
ENST00000445224.6
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr5_+_42548043 | 0.34 |
ENST00000618088.4
ENST00000612382.4 |
GHR
|
growth hormone receptor |
| chr12_+_101594849 | 0.34 |
ENST00000547405.5
ENST00000452455.6 ENST00000392934.7 ENST00000547509.5 ENST00000361685.6 ENST00000549145.5 ENST00000361466.7 ENST00000553190.5 ENST00000545503.6 ENST00000536007.5 ENST00000541119.5 ENST00000551300.5 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C1 |
| chr1_+_207325629 | 0.34 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr19_-_58353482 | 0.34 |
ENST00000263100.8
|
A1BG
|
alpha-1-B glycoprotein |
| chr16_+_86566821 | 0.33 |
ENST00000649859.1
|
FOXC2
|
forkhead box C2 |
| chr6_+_27865308 | 0.33 |
ENST00000613174.2
|
H2AC16
|
H2A clustered histone 16 |
| chr5_+_148394712 | 0.33 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr15_-_44711306 | 0.32 |
ENST00000682850.1
|
PATL2
|
PAT1 homolog 2 |
| chr5_-_88823763 | 0.32 |
ENST00000635898.1
ENST00000626391.2 ENST00000628656.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr4_-_99435396 | 0.31 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chrX_+_47585212 | 0.31 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr17_-_76141240 | 0.31 |
ENST00000322957.7
|
FOXJ1
|
forkhead box J1 |
| chr2_-_110212519 | 0.31 |
ENST00000611969.5
|
MTLN
|
mitoregulin |
| chr5_-_112419251 | 0.30 |
ENST00000261486.6
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr6_+_72212887 | 0.30 |
ENST00000523963.5
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr1_-_219613069 | 0.30 |
ENST00000367211.6
ENST00000651890.1 |
ZC3H11B
|
zinc finger CCCH-type containing 11B |
| chr7_+_1688119 | 0.30 |
ENST00000424383.4
|
ELFN1
|
extracellular leucine rich repeat and fibronectin type III domain containing 1 |
| chr1_-_17045219 | 0.30 |
ENST00000491274.5
|
SDHB
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr19_-_51417791 | 0.30 |
ENST00000353836.9
|
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr8_-_132675567 | 0.30 |
ENST00000519595.5
|
LRRC6
|
leucine rich repeat containing 6 |
| chr6_+_42050876 | 0.29 |
ENST00000465926.5
ENST00000482432.1 |
TAF8
|
TATA-box binding protein associated factor 8 |
| chr12_-_10826358 | 0.29 |
ENST00000240619.2
|
TAS2R10
|
taste 2 receptor member 10 |
| chr20_+_4686448 | 0.29 |
ENST00000379440.9
ENST00000424424.2 ENST00000457586.2 |
PRNP
|
prion protein |
| chr3_+_98147479 | 0.29 |
ENST00000641380.1
|
OR5H14
|
olfactory receptor family 5 subfamily H member 14 |
| chr15_-_19988117 | 0.29 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr13_+_108269629 | 0.27 |
ENST00000430559.5
ENST00000375887.9 |
TNFSF13B
|
TNF superfamily member 13b |
| chrX_+_55717733 | 0.26 |
ENST00000414239.5
ENST00000374941.9 |
RRAGB
|
Ras related GTP binding B |
| chrX_+_55717796 | 0.26 |
ENST00000262850.7
|
RRAGB
|
Ras related GTP binding B |
| chr8_+_12104389 | 0.26 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D |
| chr8_+_26293112 | 0.26 |
ENST00000523925.5
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr14_+_101809795 | 0.26 |
ENST00000350249.7
ENST00000557621.5 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2 regulatory subunit B'gamma |
| chr12_-_91180365 | 0.26 |
ENST00000547937.5
|
DCN
|
decorin |
| chr5_+_119476530 | 0.26 |
ENST00000645099.1
ENST00000513628.5 |
HSD17B4
|
hydroxysteroid 17-beta dehydrogenase 4 |
| chr2_+_43774033 | 0.26 |
ENST00000260605.12
ENST00000406852.7 ENST00000398823.6 ENST00000605786.5 |
DYNC2LI1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr12_-_21910853 | 0.25 |
ENST00000544039.5
|
ABCC9
|
ATP binding cassette subfamily C member 9 |
| chr6_+_26251607 | 0.25 |
ENST00000619466.2
|
H2BC9
|
H2B clustered histone 9 |
| chr20_+_5750437 | 0.25 |
ENST00000445603.1
ENST00000442185.1 |
SHLD1
|
shieldin complex subunit 1 |
| chr1_-_248277976 | 0.25 |
ENST00000641220.1
|
OR2T33
|
olfactory receptor family 2 subfamily T member 33 |
| chr11_-_125111579 | 0.25 |
ENST00000532156.5
ENST00000532407.5 ENST00000279968.8 ENST00000527766.5 ENST00000529583.5 ENST00000524373.5 ENST00000527271.5 ENST00000526175.5 ENST00000529609.5 ENST00000682305.1 ENST00000533273.1 |
TMEM218
|
transmembrane protein 218 |
| chr18_+_22933321 | 0.25 |
ENST00000327155.10
|
RBBP8
|
RB binding protein 8, endonuclease |
| chr12_-_122703346 | 0.25 |
ENST00000328880.6
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr16_+_28637654 | 0.24 |
ENST00000529716.5
|
NPIPB8
|
nuclear pore complex interacting protein family member B8 |
| chr20_+_5750381 | 0.24 |
ENST00000378979.8
ENST00000303142.11 |
SHLD1
|
shieldin complex subunit 1 |
| chr6_+_116100813 | 0.24 |
ENST00000419791.3
ENST00000319550.9 |
NT5DC1
|
5'-nucleotidase domain containing 1 |
| chr13_+_48256214 | 0.24 |
ENST00000650237.1
|
ITM2B
|
integral membrane protein 2B |
| chr3_+_148791058 | 0.24 |
ENST00000491148.5
|
CPB1
|
carboxypeptidase B1 |
| chr12_+_74537787 | 0.24 |
ENST00000519948.4
|
ATXN7L3B
|
ataxin 7 like 3B |
| chr10_+_13610047 | 0.24 |
ENST00000601460.5
|
ENSG00000282246.1
|
novel protein |
| chr3_+_159069252 | 0.23 |
ENST00000640015.1
ENST00000476809.7 ENST00000485419.7 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr9_+_122519141 | 0.23 |
ENST00000340750.1
|
OR1J4
|
olfactory receptor family 1 subfamily J member 4 |
| chr9_+_72577369 | 0.23 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr17_-_41168219 | 0.23 |
ENST00000391356.4
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr4_-_99435336 | 0.23 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr15_+_58410543 | 0.23 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr7_-_150302980 | 0.23 |
ENST00000252071.8
|
ACTR3C
|
actin related protein 3C |
| chr5_+_55160161 | 0.23 |
ENST00000296734.6
ENST00000515370.1 ENST00000503787.6 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr8_+_66775178 | 0.22 |
ENST00000396596.2
ENST00000521960.5 ENST00000522398.5 ENST00000522629.5 ENST00000520976.5 |
SGK3
|
serum/glucocorticoid regulated kinase family member 3 |
| chr4_-_67545464 | 0.22 |
ENST00000273853.11
|
CENPC
|
centromere protein C |
| chr12_-_53321544 | 0.22 |
ENST00000394384.7
ENST00000209873.9 |
AAAS
|
aladin WD repeat nucleoporin |
| chr4_-_99219230 | 0.22 |
ENST00000394897.5
ENST00000508558.1 ENST00000394899.6 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr16_+_15395745 | 0.21 |
ENST00000287594.7
ENST00000396385.4 ENST00000568766.1 |
MPV17L
ENSG00000261130.5
|
MPV17 mitochondrial inner membrane protein like novel protein |
| chr1_-_60073750 | 0.21 |
ENST00000371201.3
|
C1orf87
|
chromosome 1 open reading frame 87 |
| chr1_-_36149464 | 0.20 |
ENST00000373166.8
ENST00000373159.1 ENST00000373162.5 ENST00000616074.4 ENST00000616395.4 |
TRAPPC3
|
trafficking protein particle complex 3 |
| chr5_-_78985951 | 0.20 |
ENST00000396151.7
|
ARSB
|
arylsulfatase B |
| chr12_+_18242955 | 0.20 |
ENST00000676171.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr3_+_131026844 | 0.20 |
ENST00000510769.5
ENST00000383366.9 ENST00000510688.5 ENST00000511262.5 |
NEK11
|
NIMA related kinase 11 |
| chr3_+_119579577 | 0.20 |
ENST00000478927.5
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chrX_+_136648214 | 0.20 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr11_+_66509079 | 0.20 |
ENST00000419755.3
|
ENSG00000256349.1
|
novel protein |
| chr9_+_72577788 | 0.20 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr2_+_48568981 | 0.20 |
ENST00000394754.5
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr6_+_28349907 | 0.20 |
ENST00000252211.7
ENST00000341464.9 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr16_+_69565958 | 0.20 |
ENST00000349945.7
ENST00000354436.6 |
NFAT5
|
nuclear factor of activated T cells 5 |
| chr17_+_38003976 | 0.19 |
ENST00000616101.4
|
TBC1D3D
|
TBC1 domain family member 3D |
| chr17_-_37989048 | 0.19 |
ENST00000617678.2
ENST00000612727.5 |
TBC1D3L
|
TBC1 domain family member 3L |
| chr17_+_38127951 | 0.19 |
ENST00000621587.2
|
TBC1D3E
|
TBC1 domain family member 3E |
| chr11_-_60183011 | 0.19 |
ENST00000533023.5
ENST00000420732.6 ENST00000528851.6 |
MS4A6A
|
membrane spanning 4-domains A6A |
| chr6_+_110180418 | 0.19 |
ENST00000368930.5
ENST00000307731.2 |
CDC40
|
cell division cycle 40 |
| chr8_+_109540602 | 0.19 |
ENST00000530629.5
ENST00000620557.4 |
EBAG9
|
estrogen receptor binding site associated antigen 9 |
| chr11_+_7088991 | 0.18 |
ENST00000306904.7
|
RBMXL2
|
RBMX like 2 |
| chr21_+_30396030 | 0.18 |
ENST00000355459.4
|
KRTAP13-1
|
keratin associated protein 13-1 |
| chr14_-_89954518 | 0.18 |
ENST00000556005.1
ENST00000555872.5 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr16_-_28363508 | 0.17 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family member B6 |
| chr15_-_99249523 | 0.17 |
ENST00000560235.1
ENST00000394132.7 ENST00000560860.5 ENST00000558078.5 ENST00000560772.5 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr9_-_96778053 | 0.17 |
ENST00000375231.5
ENST00000223428.9 |
ZNF510
|
zinc finger protein 510 |
| chr6_-_29375291 | 0.17 |
ENST00000396806.3
|
OR12D3
|
olfactory receptor family 12 subfamily D member 3 |
| chr17_-_44023106 | 0.17 |
ENST00000585950.5
ENST00000592127.5 ENST00000589334.5 |
TMEM101
|
transmembrane protein 101 |
| chr1_-_34985288 | 0.17 |
ENST00000417456.1
ENST00000373337.3 |
ENSG00000284773.1
TMEM35B
|
novel protein transmembrane protein 35B |
| chr9_-_21217311 | 0.16 |
ENST00000380216.1
|
IFNA16
|
interferon alpha 16 |
| chr20_-_35411963 | 0.16 |
ENST00000349714.9
ENST00000438533.5 ENST00000359226.6 ENST00000374384.6 ENST00000374385.10 ENST00000424405.5 ENST00000397554.5 ENST00000374380.6 |
UQCC1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr3_-_151329539 | 0.16 |
ENST00000325602.6
|
P2RY13
|
purinergic receptor P2Y13 |
| chr10_+_13161543 | 0.16 |
ENST00000378714.8
ENST00000479669.5 ENST00000484800.6 |
MCM10
|
minichromosome maintenance 10 replication initiation factor |
| chr2_-_112433519 | 0.16 |
ENST00000496537.1
ENST00000330575.9 ENST00000302558.8 |
RGPD8
|
RANBP2 like and GRIP domain containing 8 |
| chr6_-_26234978 | 0.16 |
ENST00000244534.7
|
H1-3
|
H1.3 linker histone, cluster member |
| chr14_+_24070837 | 0.15 |
ENST00000537691.5
ENST00000397016.6 ENST00000560356.5 ENST00000558450.5 |
CPNE6
|
copine 6 |
| chr15_-_65133780 | 0.15 |
ENST00000204549.9
|
PDCD7
|
programmed cell death 7 |
| chr2_+_206159884 | 0.15 |
ENST00000392222.7
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chrX_+_136648138 | 0.15 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr1_+_81306096 | 0.15 |
ENST00000370721.5
ENST00000370727.5 ENST00000370725.5 ENST00000370723.5 ENST00000370728.5 ENST00000370730.5 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr10_-_69409275 | 0.15 |
ENST00000373307.5
|
TACR2
|
tachykinin receptor 2 |
| chr16_+_72054477 | 0.15 |
ENST00000355906.10
ENST00000570083.5 ENST00000228226.12 ENST00000398131.6 ENST00000569639.5 ENST00000564499.5 ENST00000357763.8 ENST00000613898.1 ENST00000562526.5 ENST00000565574.5 ENST00000568417.6 |
HP
|
haptoglobin |
| chr1_-_53940100 | 0.15 |
ENST00000371376.1
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr8_-_53842899 | 0.15 |
ENST00000524234.1
ENST00000521275.5 ENST00000396774.6 |
ATP6V1H
|
ATPase H+ transporting V1 subunit H |
| chr6_-_73521783 | 0.15 |
ENST00000331523.7
ENST00000356303.7 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr2_-_55010348 | 0.14 |
ENST00000394609.6
|
RTN4
|
reticulon 4 |
| chr1_-_36149450 | 0.14 |
ENST00000373163.5
|
TRAPPC3
|
trafficking protein particle complex 3 |
| chr7_+_90709530 | 0.14 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14 |
| chr4_-_88697810 | 0.14 |
ENST00000323061.7
|
NAP1L5
|
nucleosome assembly protein 1 like 5 |
| chr6_-_52840843 | 0.14 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr5_-_177780633 | 0.14 |
ENST00000513554.5
ENST00000440605.7 |
FAM153A
|
family with sequence similarity 153 member A |
| chr11_+_122838492 | 0.14 |
ENST00000227348.9
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 0.6 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.2 | 0.6 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.9 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 0.5 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.2 | 0.7 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.1 | 0.4 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.1 | 0.6 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.4 | GO:0071284 | copper ion export(GO:0060003) cellular response to lead ion(GO:0071284) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.7 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 1.8 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 1.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 1.5 | GO:0061141 | lung ciliated cell differentiation(GO:0061141) |
| 0.1 | 0.3 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.1 | 0.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.7 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.3 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.1 | 0.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.6 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 2.7 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.2 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.1 | 0.3 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.1 | 0.5 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.4 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 1.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.3 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.5 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.2 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 1.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 1.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 1.0 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.6 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 1.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.3 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.4 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 1.1 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 1.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 1.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.8 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.2 | GO:0042832 | carbohydrate mediated signaling(GO:0009756) defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:2000617 | positive regulation of histone H3-K9 acetylation(GO:2000617) |
| 0.0 | 0.1 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.7 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.5 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.3 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 2.0 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 3.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.0 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 1.9 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 1.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.2 | 0.7 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 1.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 0.9 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.7 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.6 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.4 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.4 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 1.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.7 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 2.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.1 | 1.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.3 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.3 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 0.6 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.2 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.0 | 1.0 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 1.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.5 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 1.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.0 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 5.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 5.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |