Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
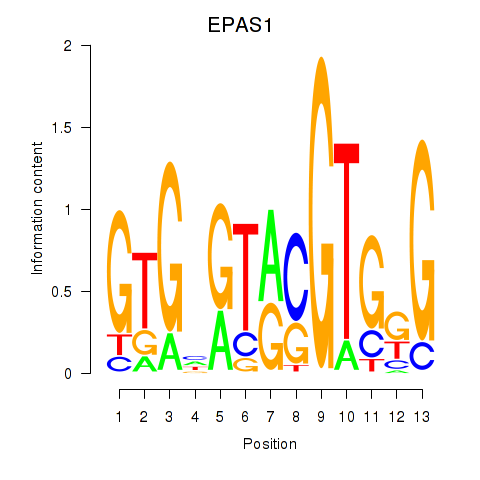
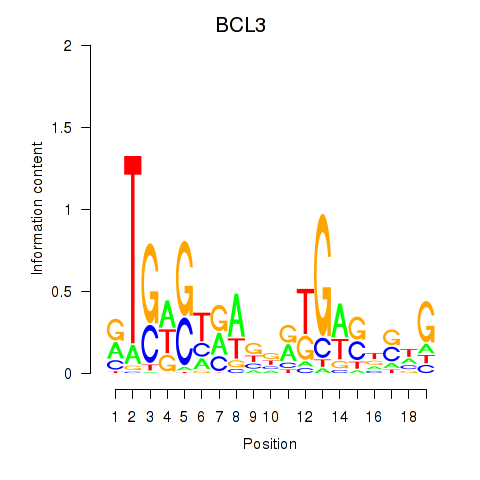
Results for EPAS1_BCL3
Z-value: 0.96
Transcription factors associated with EPAS1_BCL3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EPAS1
|
ENSG00000116016.14 | EPAS1 |
|
BCL3
|
ENSG00000069399.15 | BCL3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EPAS1 | hg38_v1_chr2_+_46297397_46297414 | -0.30 | 1.4e-01 | Click! |
| BCL3 | hg38_v1_chr19_+_44751251_44751321 | -0.18 | 4.0e-01 | Click! |
Activity profile of EPAS1_BCL3 motif
Sorted Z-values of EPAS1_BCL3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EPAS1_BCL3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_169980474 | 2.53 |
ENST00000377365.4
|
INSYN2B
|
inhibitory synaptic factor family member 2B |
| chr3_-_116444983 | 1.42 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr14_+_32934383 | 1.35 |
ENST00000551634.6
|
NPAS3
|
neuronal PAS domain protein 3 |
| chr16_+_57628507 | 1.24 |
ENST00000456916.5
ENST00000567154.5 ENST00000388813.9 ENST00000562558.6 ENST00000566271.6 ENST00000563374.5 ENST00000673126.2 ENST00000562631.7 ENST00000568234.5 ENST00000565770.5 ENST00000566164.5 |
ADGRG1
|
adhesion G protein-coupled receptor G1 |
| chr7_+_154305256 | 1.22 |
ENST00000619756.4
|
DPP6
|
dipeptidyl peptidase like 6 |
| chr17_-_43545636 | 0.98 |
ENST00000393664.6
|
ETV4
|
ETS variant transcription factor 4 |
| chr8_+_62248591 | 0.88 |
ENST00000519049.6
|
NKAIN3
|
sodium/potassium transporting ATPase interacting 3 |
| chr1_-_44031446 | 0.82 |
ENST00000372310.8
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr1_+_40374648 | 0.81 |
ENST00000372708.5
|
SMAP2
|
small ArfGAP2 |
| chr1_-_150235972 | 0.80 |
ENST00000534220.1
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr11_-_72722302 | 0.75 |
ENST00000334211.12
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr17_-_43546323 | 0.71 |
ENST00000545954.5
ENST00000319349.10 |
ETV4
|
ETS variant transcription factor 4 |
| chr7_-_27152561 | 0.70 |
ENST00000467897.2
ENST00000612286.5 ENST00000498652.1 |
ENSG00000273433.1
HOXA3
ENSG00000272801.1
|
novel transcript homeobox A3 novel transcript |
| chr17_-_43545707 | 0.70 |
ENST00000545089.5
|
ETV4
|
ETS variant transcription factor 4 |
| chr5_-_136365476 | 0.69 |
ENST00000378459.7
ENST00000502753.4 ENST00000513104.6 ENST00000352189.8 |
TRPC7
|
transient receptor potential cation channel subfamily C member 7 |
| chr1_+_115641945 | 0.68 |
ENST00000355485.7
ENST00000369510.8 |
VANGL1
|
VANGL planar cell polarity protein 1 |
| chr12_+_65824475 | 0.68 |
ENST00000403681.7
|
HMGA2
|
high mobility group AT-hook 2 |
| chr8_-_69834970 | 0.66 |
ENST00000260126.9
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr6_-_42722590 | 0.66 |
ENST00000230381.7
|
PRPH2
|
peripherin 2 |
| chr22_-_37149900 | 0.64 |
ENST00000216223.10
|
IL2RB
|
interleukin 2 receptor subunit beta |
| chr1_-_44031352 | 0.63 |
ENST00000372306.7
ENST00000475075.6 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr9_+_72628020 | 0.59 |
ENST00000646619.1
|
TMC1
|
transmembrane channel like 1 |
| chr8_+_76683779 | 0.57 |
ENST00000523885.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr14_+_70641896 | 0.53 |
ENST00000256367.3
|
TTC9
|
tetratricopeptide repeat domain 9 |
| chr19_+_55857437 | 0.53 |
ENST00000587891.5
|
NLRP4
|
NLR family pyrin domain containing 4 |
| chr17_-_43545891 | 0.52 |
ENST00000591713.5
ENST00000538265.5 |
ETV4
|
ETS variant transcription factor 4 |
| chr3_-_58587033 | 0.50 |
ENST00000447756.2
|
FAM107A
|
family with sequence similarity 107 member A |
| chr1_+_184386978 | 0.50 |
ENST00000235307.7
|
C1orf21
|
chromosome 1 open reading frame 21 |
| chr22_+_41976933 | 0.49 |
ENST00000396425.7
|
SEPTIN3
|
septin 3 |
| chr2_-_168913277 | 0.48 |
ENST00000451987.5
|
SPC25
|
SPC25 component of NDC80 kinetochore complex |
| chr7_-_41703062 | 0.48 |
ENST00000242208.5
|
INHBA
|
inhibin subunit beta A |
| chr2_+_205683109 | 0.48 |
ENST00000357118.8
ENST00000272849.7 ENST00000412873.2 |
NRP2
|
neuropilin 2 |
| chr14_+_21887848 | 0.46 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chr3_-_9249623 | 0.46 |
ENST00000383836.8
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr19_+_41219235 | 0.46 |
ENST00000359092.7
|
AXL
|
AXL receptor tyrosine kinase |
| chr19_+_15049469 | 0.45 |
ENST00000427043.4
|
CASP14
|
caspase 14 |
| chr11_-_75351686 | 0.44 |
ENST00000360025.7
|
ARRB1
|
arrestin beta 1 |
| chr12_-_16608183 | 0.43 |
ENST00000354662.5
ENST00000538051.5 |
LMO3
|
LIM domain only 3 |
| chr17_+_4715438 | 0.43 |
ENST00000571206.1
|
ARRB2
|
arrestin beta 2 |
| chr13_-_33205997 | 0.42 |
ENST00000399365.7
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr7_+_87628538 | 0.42 |
ENST00000394654.3
|
RUNDC3B
|
RUN domain containing 3B |
| chr2_+_74654228 | 0.42 |
ENST00000611975.4
ENST00000357877.7 ENST00000339773.9 ENST00000434486.5 |
SEMA4F
|
ssemaphorin 4F |
| chr5_-_59356962 | 0.42 |
ENST00000405755.6
|
PDE4D
|
phosphodiesterase 4D |
| chr9_+_92974476 | 0.42 |
ENST00000337352.10
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr3_-_127823235 | 0.41 |
ENST00000398104.5
|
MGLL
|
monoglyceride lipase |
| chr19_+_58326991 | 0.41 |
ENST00000329665.5
|
ZSCAN22
|
zinc finger and SCAN domain containing 22 |
| chr22_+_39456996 | 0.41 |
ENST00000341184.7
|
MGAT3
|
beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase |
| chr19_-_48513161 | 0.41 |
ENST00000673139.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr2_-_100142575 | 0.40 |
ENST00000317233.8
ENST00000672204.1 ENST00000416492.5 ENST00000672857.1 ENST00000672756.2 |
AFF3
|
AF4/FMR2 family member 3 |
| chr12_-_117968972 | 0.40 |
ENST00000339824.7
|
KSR2
|
kinase suppressor of ras 2 |
| chr3_-_127823177 | 0.40 |
ENST00000434178.6
|
MGLL
|
monoglyceride lipase |
| chr9_+_72577939 | 0.40 |
ENST00000645773.1
|
TMC1
|
transmembrane channel like 1 |
| chr3_-_127822835 | 0.39 |
ENST00000453507.6
|
MGLL
|
monoglyceride lipase |
| chr9_+_72577788 | 0.39 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr7_-_117323041 | 0.38 |
ENST00000491214.1
ENST00000265441.8 |
WNT2
|
Wnt family member 2 |
| chr3_+_14402592 | 0.38 |
ENST00000622186.5
ENST00000621751.4 |
SLC6A6
|
solute carrier family 6 member 6 |
| chr19_+_44942268 | 0.38 |
ENST00000591600.1
|
APOC4
|
apolipoprotein C4 |
| chr12_-_2835018 | 0.37 |
ENST00000337508.9
|
NRIP2
|
nuclear receptor interacting protein 2 |
| chr5_-_59039454 | 0.37 |
ENST00000358923.10
|
PDE4D
|
phosphodiesterase 4D |
| chr12_-_16608073 | 0.37 |
ENST00000441439.6
|
LMO3
|
LIM domain only 3 |
| chr1_-_30757767 | 0.37 |
ENST00000294507.4
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr19_-_35134861 | 0.36 |
ENST00000591633.2
|
LGI4
|
leucine rich repeat LGI family member 4 |
| chr13_-_101416441 | 0.36 |
ENST00000675332.1
ENST00000676315.1 ENST00000251127.11 |
NALCN
|
sodium leak channel, non-selective |
| chr13_-_52739769 | 0.35 |
ENST00000448904.6
ENST00000377962.8 |
CNMD
|
chondromodulin |
| chr9_+_2621766 | 0.35 |
ENST00000382100.8
|
VLDLR
|
very low density lipoprotein receptor |
| chr8_-_27479229 | 0.34 |
ENST00000407991.3
|
CHRNA2
|
cholinergic receptor nicotinic alpha 2 subunit |
| chr12_+_26195647 | 0.34 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr5_+_52989314 | 0.34 |
ENST00000296585.10
|
ITGA2
|
integrin subunit alpha 2 |
| chr22_+_41976760 | 0.33 |
ENST00000396426.7
ENST00000406029.5 |
SEPTIN3
|
septin 3 |
| chr9_+_2622053 | 0.33 |
ENST00000681306.1
ENST00000681618.1 |
VLDLR
|
very low density lipoprotein receptor |
| chr7_+_87628355 | 0.33 |
ENST00000338056.7
ENST00000493037.5 |
RUNDC3B
|
RUN domain containing 3B |
| chr3_-_185152974 | 0.33 |
ENST00000335012.3
|
C3orf70
|
chromosome 3 open reading frame 70 |
| chr17_-_8087709 | 0.33 |
ENST00000647874.1
|
ALOX12B
|
arachidonate 12-lipoxygenase, 12R type |
| chr11_+_128891468 | 0.33 |
ENST00000338350.4
|
KCNJ5
|
potassium inwardly rectifying channel subfamily J member 5 |
| chr14_+_22226711 | 0.32 |
ENST00000390463.3
|
TRAV36DV7
|
T cell receptor alpha variable 36/delta variable 7 |
| chr3_-_58627596 | 0.32 |
ENST00000474531.5
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107 member A |
| chr15_+_33310946 | 0.32 |
ENST00000415757.7
ENST00000634891.2 ENST00000389232.9 ENST00000622037.1 |
RYR3
|
ryanodine receptor 3 |
| chr19_+_10452891 | 0.32 |
ENST00000344979.7
|
PDE4A
|
phosphodiesterase 4A |
| chr5_-_173328407 | 0.32 |
ENST00000265087.9
|
STC2
|
stanniocalcin 2 |
| chr7_-_38330935 | 0.32 |
ENST00000390343.2
|
TRGV8
|
T cell receptor gamma variable 8 |
| chrX_-_20116871 | 0.32 |
ENST00000379651.7
ENST00000443379.7 ENST00000379643.10 |
MAP7D2
|
MAP7 domain containing 2 |
| chr6_-_31652358 | 0.32 |
ENST00000456622.5
ENST00000435080.5 ENST00000375976.8 ENST00000441054.5 |
BAG6
|
BAG cochaperone 6 |
| chr8_+_62248933 | 0.31 |
ENST00000523211.5
ENST00000524201.1 |
NKAIN3
|
sodium/potassium transporting ATPase interacting 3 |
| chr9_+_2621556 | 0.31 |
ENST00000680746.1
|
VLDLR
|
very low density lipoprotein receptor |
| chr9_+_18474100 | 0.31 |
ENST00000327883.11
ENST00000431052.6 ENST00000380570.8 ENST00000380548.9 |
ADAMTSL1
|
ADAMTS like 1 |
| chr3_-_128493173 | 0.31 |
ENST00000498200.1
ENST00000341105.7 |
GATA2
|
GATA binding protein 2 |
| chr2_-_85418421 | 0.31 |
ENST00000409275.1
|
CAPG
|
capping actin protein, gelsolin like |
| chr11_+_10450627 | 0.31 |
ENST00000396554.7
ENST00000524866.5 |
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr19_-_11197516 | 0.31 |
ENST00000592903.5
ENST00000586659.6 ENST00000589359.5 ENST00000588724.5 |
KANK2
|
KN motif and ankyrin repeat domains 2 |
| chr6_-_11807045 | 0.30 |
ENST00000379415.6
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr7_-_100167222 | 0.30 |
ENST00000411994.1
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4 |
| chr7_+_20615653 | 0.30 |
ENST00000404938.7
|
ABCB5
|
ATP binding cassette subfamily B member 5 |
| chr1_-_153565535 | 0.30 |
ENST00000368707.5
|
S100A2
|
S100 calcium binding protein A2 |
| chr19_-_51387955 | 0.30 |
ENST00000221973.7
ENST00000596399.2 |
LIM2
|
lens intrinsic membrane protein 2 |
| chr14_+_79279403 | 0.30 |
ENST00000281127.11
|
NRXN3
|
neurexin 3 |
| chr12_-_24562438 | 0.30 |
ENST00000646273.1
ENST00000659413.1 ENST00000446891.7 |
SOX5
|
SRY-box transcription factor 5 |
| chr3_-_158105718 | 0.29 |
ENST00000554685.2
|
SHOX2
|
short stature homeobox 2 |
| chr2_+_218382265 | 0.29 |
ENST00000233202.11
|
SLC11A1
|
solute carrier family 11 member 1 |
| chr6_+_43770202 | 0.29 |
ENST00000372067.8
ENST00000672860.2 |
VEGFA
|
vascular endothelial growth factor A |
| chr11_-_134411854 | 0.29 |
ENST00000392580.5
|
B3GAT1
|
beta-1,3-glucuronyltransferase 1 |
| chr1_+_13584262 | 0.29 |
ENST00000376061.8
ENST00000513143.5 |
PDPN
|
podoplanin |
| chr11_-_69819410 | 0.29 |
ENST00000334134.4
|
FGF3
|
fibroblast growth factor 3 |
| chr4_-_121072519 | 0.29 |
ENST00000379692.9
|
NDNF
|
neuron derived neurotrophic factor |
| chr11_-_79441016 | 0.29 |
ENST00000278550.12
|
TENM4
|
teneurin transmembrane protein 4 |
| chr16_+_83971451 | 0.29 |
ENST00000565691.5
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr20_+_52972347 | 0.28 |
ENST00000371497.10
|
TSHZ2
|
teashirt zinc finger homeobox 2 |
| chr7_-_130686545 | 0.28 |
ENST00000356588.8
ENST00000443954.5 ENST00000438346.1 |
TSGA13
|
testis specific 13 |
| chr8_+_109086585 | 0.28 |
ENST00000518632.2
|
TRHR
|
thyrotropin releasing hormone receptor |
| chr15_+_48191648 | 0.28 |
ENST00000646012.1
ENST00000561127.5 ENST00000647546.1 ENST00000559641.5 ENST00000417307.3 |
SLC12A1
CTXN2
|
solute carrier family 12 member 1 cortexin 2 |
| chr17_+_21376321 | 0.28 |
ENST00000583088.6
|
KCNJ12
|
potassium inwardly rectifying channel subfamily J member 12 |
| chr11_+_76783349 | 0.28 |
ENST00000333090.5
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr7_+_75398915 | 0.28 |
ENST00000437796.1
|
TRIM73
|
tripartite motif containing 73 |
| chr17_-_31321603 | 0.28 |
ENST00000462804.3
|
EVI2A
|
ecotropic viral integration site 2A |
| chrX_+_1268786 | 0.27 |
ENST00000501036.7
ENST00000417535.7 |
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha |
| chr14_+_93927358 | 0.27 |
ENST00000557000.2
|
FAM181A
|
family with sequence similarity 181 member A |
| chr7_+_142332182 | 0.27 |
ENST00000547918.2
|
TRBV7-1
|
T cell receptor beta variable 7-1 (non-functional) |
| chr14_-_91244669 | 0.27 |
ENST00000650645.1
|
GPR68
|
G protein-coupled receptor 68 |
| chr14_-_91244508 | 0.27 |
ENST00000535815.5
ENST00000529102.1 |
GPR68
|
G protein-coupled receptor 68 |
| chr5_+_175872741 | 0.27 |
ENST00000502265.5
|
CPLX2
|
complexin 2 |
| chr12_-_102481744 | 0.27 |
ENST00000644491.1
|
IGF1
|
insulin like growth factor 1 |
| chr11_-_30586866 | 0.27 |
ENST00000528686.2
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chrX_+_1268807 | 0.27 |
ENST00000381524.8
ENST00000381529.9 ENST00000412290.6 |
CSF2RA
|
colony stimulating factor 2 receptor subunit alpha |
| chr1_-_44017296 | 0.27 |
ENST00000357730.6
ENST00000360584.6 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr22_+_21632751 | 0.26 |
ENST00000292779.4
|
CCDC116
|
coiled-coil domain containing 116 |
| chr2_+_241687059 | 0.26 |
ENST00000636051.1
|
ING5
|
inhibitor of growth family member 5 |
| chr19_+_41219177 | 0.26 |
ENST00000301178.9
|
AXL
|
AXL receptor tyrosine kinase |
| chr12_+_6914571 | 0.26 |
ENST00000229277.6
ENST00000538763.5 ENST00000545045.6 |
ENO2
|
enolase 2 |
| chr19_+_39704461 | 0.26 |
ENST00000360675.7
ENST00000392052.8 ENST00000601802.1 |
LGALS14
|
galectin 14 |
| chr3_-_127822455 | 0.26 |
ENST00000265052.10
|
MGLL
|
monoglyceride lipase |
| chr7_+_114414997 | 0.26 |
ENST00000462331.5
ENST00000393491.7 ENST00000403559.8 ENST00000408937.7 ENST00000393498.6 ENST00000393495.7 ENST00000378237.7 |
FOXP2
|
forkhead box P2 |
| chr20_+_9069076 | 0.26 |
ENST00000378473.9
|
PLCB4
|
phospholipase C beta 4 |
| chr16_-_30010972 | 0.26 |
ENST00000565273.5
ENST00000567332.6 ENST00000350119.9 |
DOC2A
|
double C2 domain alpha |
| chr19_-_43082692 | 0.26 |
ENST00000406487.6
|
PSG2
|
pregnancy specific beta-1-glycoprotein 2 |
| chr7_-_36724380 | 0.25 |
ENST00000617267.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr19_-_42917837 | 0.25 |
ENST00000292125.6
ENST00000402603.8 ENST00000594375.1 ENST00000187910.7 |
PSG6
|
pregnancy specific beta-1-glycoprotein 6 |
| chr4_+_61202142 | 0.25 |
ENST00000514591.5
|
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr11_+_105610883 | 0.25 |
ENST00000531011.5
ENST00000525187.5 ENST00000530497.1 |
GRIA4
|
glutamate ionotropic receptor AMPA type subunit 4 |
| chr18_+_46334197 | 0.25 |
ENST00000588679.1
ENST00000543885.2 |
RNF165
|
ring finger protein 165 |
| chr13_-_101416362 | 0.24 |
ENST00000675802.1
ENST00000675150.1 |
NALCN
|
sodium leak channel, non-selective |
| chr6_+_34236865 | 0.24 |
ENST00000674029.1
ENST00000447654.5 ENST00000347617.10 ENST00000401473.7 ENST00000311487.9 |
HMGA1
|
high mobility group AT-hook 1 |
| chrX_-_11265975 | 0.24 |
ENST00000303025.10
ENST00000657361.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr14_+_95876385 | 0.24 |
ENST00000504119.1
|
TUNAR
|
TCL1 upstream neural differentiation-associated RNA |
| chr19_+_12995467 | 0.24 |
ENST00000592199.6
|
NFIX
|
nuclear factor I X |
| chr22_+_31092447 | 0.23 |
ENST00000455608.5
|
SMTN
|
smoothelin |
| chr3_+_14402576 | 0.23 |
ENST00000613060.4
|
SLC6A6
|
solute carrier family 6 member 6 |
| chr7_+_114414809 | 0.23 |
ENST00000350908.9
|
FOXP2
|
forkhead box P2 |
| chr3_-_179072205 | 0.23 |
ENST00000432729.5
|
ZMAT3
|
zinc finger matrin-type 3 |
| chr7_+_127593727 | 0.23 |
ENST00000478821.1
ENST00000265825.6 |
FSCN3
|
fascin actin-bundling protein 3 |
| chr19_-_16471943 | 0.23 |
ENST00000602151.1
ENST00000597937.5 ENST00000455140.7 ENST00000535753.6 |
EPS15L1
|
epidermal growth factor receptor pathway substrate 15 like 1 |
| chr11_-_27700472 | 0.23 |
ENST00000418212.5
ENST00000533246.5 |
BDNF
|
brain derived neurotrophic factor |
| chr5_+_80960354 | 0.23 |
ENST00000265080.9
|
RASGRF2
|
Ras protein specific guanine nucleotide releasing factor 2 |
| chr6_-_24358036 | 0.23 |
ENST00000378454.8
|
DCDC2
|
doublecortin domain containing 2 |
| chrX_-_143635081 | 0.23 |
ENST00000338017.8
|
SLITRK4
|
SLIT and NTRK like family member 4 |
| chr1_+_22643626 | 0.23 |
ENST00000374640.9
ENST00000374639.7 ENST00000374637.1 |
C1QC
|
complement C1q C chain |
| chr5_-_181205284 | 0.23 |
ENST00000334421.5
|
TRIM7
|
tripartite motif containing 7 |
| chr7_-_27102669 | 0.23 |
ENST00000222718.7
|
HOXA2
|
homeobox A2 |
| chr5_+_32711313 | 0.23 |
ENST00000265074.13
|
NPR3
|
natriuretic peptide receptor 3 |
| chrX_-_72305892 | 0.23 |
ENST00000450875.5
ENST00000417400.1 ENST00000431381.5 ENST00000445983.5 ENST00000651998.1 |
CITED1
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 1 |
| chr2_-_40452046 | 0.22 |
ENST00000406785.6
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr12_-_10884244 | 0.22 |
ENST00000543626.4
|
PRH1
|
proline rich protein HaeIII subfamily 1 |
| chr22_-_37427433 | 0.22 |
ENST00000452946.1
ENST00000402918.7 |
ELFN2
ELFN2
|
extracellular leucine rich repeat and fibronectin type III domain containing 2 extracellular leucine rich repeat and fibronectin type III domain containing 2 |
| chr11_+_31812307 | 0.22 |
ENST00000643436.1
ENST00000646959.1 ENST00000645942.1 ENST00000530348.5 |
PAUPAR
ENSG00000285283.1
|
PAX6 upstream antisense RNA novel protein |
| chr2_+_219572304 | 0.22 |
ENST00000243786.3
|
INHA
|
inhibin subunit alpha |
| chr6_+_36678699 | 0.22 |
ENST00000405375.5
ENST00000244741.10 ENST00000373711.3 |
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr4_-_167234266 | 0.22 |
ENST00000511269.5
ENST00000506697.5 ENST00000512042.1 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr11_-_75351609 | 0.22 |
ENST00000420843.7
|
ARRB1
|
arrestin beta 1 |
| chr21_-_30497160 | 0.22 |
ENST00000334058.3
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr22_-_21551461 | 0.22 |
ENST00000433039.2
|
RIMBP3C
|
RIMS binding protein 3C |
| chr19_-_39320839 | 0.22 |
ENST00000248668.5
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr12_-_54984667 | 0.22 |
ENST00000524668.5
ENST00000533607.1 ENST00000449076.6 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr19_+_30372364 | 0.22 |
ENST00000355537.4
|
ZNF536
|
zinc finger protein 536 |
| chr14_+_104581141 | 0.21 |
ENST00000410013.1
|
C14orf180
|
chromosome 14 open reading frame 180 |
| chr11_-_8268716 | 0.21 |
ENST00000428101.6
|
LMO1
|
LIM domain only 1 |
| chr11_+_10450289 | 0.21 |
ENST00000444303.6
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr5_-_170389349 | 0.21 |
ENST00000274629.9
|
KCNMB1
|
potassium calcium-activated channel subfamily M regulatory beta subunit 1 |
| chr22_-_18611919 | 0.21 |
ENST00000619918.1
|
RIMBP3
|
RIMS binding protein 3 |
| chr11_+_34642468 | 0.21 |
ENST00000527935.1
|
EHF
|
ETS homologous factor |
| chr14_+_22052503 | 0.21 |
ENST00000390449.3
|
TRAV21
|
T cell receptor alpha variable 21 |
| chr11_+_117986386 | 0.21 |
ENST00000227752.8
|
IL10RA
|
interleukin 10 receptor subunit alpha |
| chr10_+_92593112 | 0.21 |
ENST00000260731.5
|
KIF11
|
kinesin family member 11 |
| chr18_+_34493289 | 0.21 |
ENST00000682923.1
ENST00000596745.5 ENST00000283365.14 ENST00000315456.10 ENST00000598774.6 ENST00000684266.1 ENST00000683092.1 ENST00000683379.1 ENST00000684359.1 |
DTNA
|
dystrobrevin alpha |
| chr4_+_85827891 | 0.21 |
ENST00000514229.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr17_+_6070361 | 0.21 |
ENST00000317744.10
|
WSCD1
|
WSC domain containing 1 |
| chr17_-_58328756 | 0.20 |
ENST00000268893.10
ENST00000343736.9 |
TSPOAP1
|
TSPO associated protein 1 |
| chr6_-_41286665 | 0.20 |
ENST00000589614.5
ENST00000244709.9 ENST00000334475.10 ENST00000591620.1 |
TREM1
|
triggering receptor expressed on myeloid cells 1 |
| chr19_+_44942230 | 0.20 |
ENST00000592954.2
ENST00000589057.5 |
APOC4
APOC4-APOC2
|
apolipoprotein C4 APOC4-APOC2 readthrough (NMD candidate) |
| chr18_+_65751000 | 0.20 |
ENST00000397968.4
|
CDH7
|
cadherin 7 |
| chr4_+_85827745 | 0.20 |
ENST00000509300.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_+_127629161 | 0.20 |
ENST00000342480.7
|
PODXL2
|
podocalyxin like 2 |
| chr22_+_31127749 | 0.20 |
ENST00000402238.5
ENST00000404453.5 ENST00000401755.1 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr6_-_31592952 | 0.20 |
ENST00000376073.8
ENST00000376072.7 |
NCR3
|
natural cytotoxicity triggering receptor 3 |
| chr1_-_247758680 | 0.20 |
ENST00000408896.4
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1 |
| chrX_+_17375230 | 0.20 |
ENST00000380060.7
|
NHS
|
NHS actin remodeling regulator |
| chr17_-_31314066 | 0.20 |
ENST00000577894.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr8_-_27478851 | 0.20 |
ENST00000240132.7
|
CHRNA2
|
cholinergic receptor nicotinic alpha 2 subunit |
| chr2_-_96035974 | 0.20 |
ENST00000434632.5
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr1_-_211492111 | 0.20 |
ENST00000367002.5
ENST00000680073.1 |
RD3
|
RD3 regulator of GUCY2D |
| chr13_-_78603539 | 0.20 |
ENST00000377208.7
|
POU4F1
|
POU class 4 homeobox 1 |
| chr4_+_89111521 | 0.20 |
ENST00000603357.3
|
TIGD2
|
tigger transposable element derived 2 |
| chr12_+_56021316 | 0.20 |
ENST00000547791.2
|
IKZF4
|
IKAROS family zinc finger 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 1.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.2 | 0.7 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.2 | 0.5 | GO:0021757 | caudate nucleus development(GO:0021757) putamen development(GO:0021758) |
| 0.2 | 1.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.6 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 1.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 1.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.5 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 1.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.4 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.1 | 0.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.1 | 0.3 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.7 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.3 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.3 | GO:0048058 | compound eye corneal lens development(GO:0048058) |
| 0.1 | 0.2 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 0.6 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.3 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.2 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 0.8 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.4 | GO:1902943 | regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) |
| 0.1 | 0.3 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.8 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.3 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.1 | 0.2 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.2 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 | 0.2 | GO:0021593 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 0.3 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.3 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.1 | 0.3 | GO:1904021 | negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.1 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 1.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.4 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.7 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.1 | 0.2 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.3 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.6 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.1 | 0.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.2 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.1 | 0.5 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.2 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.1 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.6 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.2 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.3 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.1 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.1 | 0.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.3 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 | 0.7 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.7 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.0 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.4 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.7 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.3 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.4 | GO:0097490 | sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 1.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.4 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 | 0.4 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.3 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.0 | 1.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0052148 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.3 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.4 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.1 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.1 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.2 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.4 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0035744 | T-helper 1 cell cytokine production(GO:0035744) |
| 0.0 | 0.3 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:0090133 | corticotropin hormone secreting cell differentiation(GO:0060128) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.2 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.1 | GO:1990918 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.2 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.2 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.4 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.2 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 0.0 | 0.1 | GO:1903182 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.0 | 0.1 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.0 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.4 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.2 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.4 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:0098923 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.3 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0051792 | short-chain fatty acid biosynthetic process(GO:0051790) medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.0 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.0 | GO:0030035 | microspike assembly(GO:0030035) actin filament uncapping(GO:0051695) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.0 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.5 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.2 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0006029 | proteoglycan metabolic process(GO:0006029) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.9 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.0 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.0 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.2 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.3 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 1.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 1.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.5 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 0.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 1.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.3 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.4 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.7 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 1.1 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 1.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.2 | 0.7 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.2 | 0.4 | GO:0031691 | alpha-1A adrenergic receptor binding(GO:0031691) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.2 | 0.8 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.2 | 0.5 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.2 | 0.2 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.2 | 0.6 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.1 | 1.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.3 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.1 | 0.2 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.4 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.3 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.1 | 0.3 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 1.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 1.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0042835 | BRE binding(GO:0042835) |
| 0.1 | 0.3 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.1 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.4 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 1.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.2 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.3 | GO:0098639 | collagen receptor activity(GO:0038064) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.9 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.9 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.1 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 1.0 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 1.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.0 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.0 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.0 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.0 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 1.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.8 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |