Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for ESR1
Z-value: 0.94
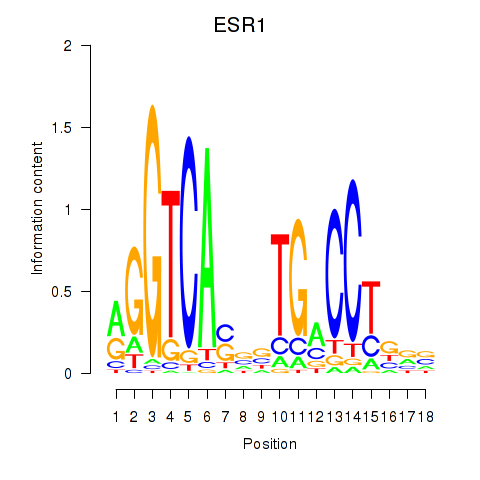
Transcription factors associated with ESR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESR1
|
ENSG00000091831.24 | ESR1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESR1 | hg38_v1_chr6_+_151807674_151807692 | 0.10 | 6.2e-01 | Click! |
Activity profile of ESR1 motif
Sorted Z-values of ESR1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ESR1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_42483604 | 2.06 |
ENST00000640132.1
|
PRICKLE1
|
prickle planar cell polarity protein 1 |
| chr12_-_42483958 | 0.99 |
ENST00000548696.6
ENST00000552240.6 |
PRICKLE1
|
prickle planar cell polarity protein 1 |
| chr19_-_1513189 | 0.99 |
ENST00000395467.6
|
ADAMTSL5
|
ADAMTS like 5 |
| chr19_+_14072754 | 0.84 |
ENST00000587086.2
|
MISP3
|
MISP family member 3 |
| chr9_+_123356189 | 0.81 |
ENST00000373631.8
|
CRB2
|
crumbs cell polarity complex component 2 |
| chr17_-_36176636 | 0.81 |
ENST00000611257.5
ENST00000616006.1 |
TBC1D3B
|
TBC1 domain family member 3B |
| chr19_-_45322867 | 0.79 |
ENST00000221476.4
|
CKM
|
creatine kinase, M-type |
| chr10_+_52314272 | 0.73 |
ENST00000373970.4
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr16_-_75266781 | 0.72 |
ENST00000420641.7
|
BCAR1
|
BCAR1 scaffold protein, Cas family member |
| chr19_-_1513003 | 0.69 |
ENST00000330475.9
ENST00000586272.5 ENST00000590562.5 |
ADAMTSL5
|
ADAMTS like 5 |
| chr10_+_110497898 | 0.68 |
ENST00000369583.4
|
DUSP5
|
dual specificity phosphatase 5 |
| chr11_-_35419462 | 0.64 |
ENST00000643522.1
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr6_+_32154131 | 0.60 |
ENST00000375143.6
ENST00000324816.11 ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr12_+_122203740 | 0.60 |
ENST00000537991.1
|
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr6_+_32154010 | 0.59 |
ENST00000375137.6
|
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr11_-_35419213 | 0.57 |
ENST00000642171.1
ENST00000644050.1 ENST00000643134.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr17_-_36264514 | 0.56 |
ENST00000618620.4
ENST00000621034.1 |
TBC1D3I
|
TBC1 domain family member 3I |
| chr19_+_50415799 | 0.55 |
ENST00000599632.1
|
ENSG00000142539.9
|
novel protein |
| chr9_-_124500986 | 0.55 |
ENST00000373587.3
|
NR5A1
|
nuclear receptor subfamily 5 group A member 1 |
| chr17_-_36388423 | 0.53 |
ENST00000610350.3
|
TBC1D3H
|
TBC1 domain family member 3H |
| chr17_+_37924415 | 0.53 |
ENST00000620247.1
|
TBC1D3K
|
TBC1 domain family member 3K |
| chr1_+_37474572 | 0.53 |
ENST00000373087.7
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr17_-_38192555 | 0.53 |
ENST00000620215.3
|
TBC1D3
|
TBC1 domain family member 3 |
| chr17_-_38068634 | 0.52 |
ENST00000622206.2
|
TBC1D3C
|
TBC1 domain family member 3C |
| chr4_-_10021490 | 0.52 |
ENST00000264784.8
|
SLC2A9
|
solute carrier family 2 member 9 |
| chr12_-_94650506 | 0.51 |
ENST00000261226.9
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr17_-_35880350 | 0.51 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr20_+_1266263 | 0.50 |
ENST00000649598.1
ENST00000381867.6 ENST00000381873.7 |
SNPH
|
syntaphilin |
| chr8_+_62248591 | 0.49 |
ENST00000519049.6
|
NKAIN3
|
sodium/potassium transporting ATPase interacting 3 |
| chr22_+_22030934 | 0.48 |
ENST00000390282.2
|
IGLV4-69
|
immunoglobulin lambda variable 4-69 |
| chr19_-_13102848 | 0.48 |
ENST00000264824.5
|
LYL1
|
LYL1 basic helix-loop-helix family member |
| chr17_+_38003976 | 0.48 |
ENST00000616101.4
|
TBC1D3D
|
TBC1 domain family member 3D |
| chr11_-_35420050 | 0.48 |
ENST00000395753.6
ENST00000395750.6 ENST00000645634.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr5_+_321695 | 0.47 |
ENST00000684583.1
ENST00000514523.1 |
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr17_+_38127951 | 0.47 |
ENST00000621587.2
|
TBC1D3E
|
TBC1 domain family member 3E |
| chr17_+_36323884 | 0.47 |
ENST00000569055.2
|
TBC1D3G
|
TBC1 domain family member 3G |
| chr17_-_36439566 | 0.47 |
ENST00000620210.1
|
TBC1D3F
|
TBC1 domain family member 3F |
| chr6_-_32154326 | 0.47 |
ENST00000475826.1
ENST00000485392.5 ENST00000494332.5 ENST00000498575.1 ENST00000428778.5 |
ENSG00000284954.1
ENSG00000285085.1
|
novel transcript novel protein |
| chr10_+_102245371 | 0.46 |
ENST00000676513.1
ENST00000676939.1 ENST00000677947.1 ENST00000677247.1 ENST00000369983.4 ENST00000678351.1 ENST00000679238.1 ENST00000677439.1 ENST00000677240.1 ENST00000677618.1 ENST00000673650.1 ENST00000674034.1 ENST00000676993.1 |
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr6_+_32153441 | 0.45 |
ENST00000414204.5
ENST00000361568.6 ENST00000395523.5 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr19_-_18884219 | 0.45 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr11_+_57542641 | 0.45 |
ENST00000527972.5
ENST00000399154.3 |
SMTNL1
|
smoothelin like 1 |
| chr17_-_37989048 | 0.45 |
ENST00000617678.2
ENST00000612727.5 |
TBC1D3L
|
TBC1 domain family member 3L |
| chr3_+_37975773 | 0.44 |
ENST00000436654.1
|
CTDSPL
|
CTD small phosphatase like |
| chr2_-_70995336 | 0.44 |
ENST00000606025.5
|
ENSG00000258881.6
|
novel protein |
| chr11_-_35420017 | 0.43 |
ENST00000643000.1
ENST00000646099.1 ENST00000647372.1 ENST00000642578.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr19_+_48872412 | 0.43 |
ENST00000200453.6
|
PPP1R15A
|
protein phosphatase 1 regulatory subunit 15A |
| chrX_+_47078330 | 0.42 |
ENST00000457380.5
|
RGN
|
regucalcin |
| chrX_+_47078380 | 0.42 |
ENST00000352078.8
|
RGN
|
regucalcin |
| chr1_-_155188351 | 0.42 |
ENST00000462317.5
|
MUC1
|
mucin 1, cell surface associated |
| chrY_+_9487313 | 0.42 |
ENST00000440215.4
|
TSPY9P
|
testis specific protein Y-linked 9, pseudogene |
| chr17_+_44847905 | 0.40 |
ENST00000587021.1
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
| chr11_-_27700447 | 0.40 |
ENST00000356660.9
|
BDNF
|
brain derived neurotrophic factor |
| chr19_-_6416773 | 0.39 |
ENST00000595223.5
|
KHSRP
|
KH-type splicing regulatory protein |
| chr1_+_206897435 | 0.39 |
ENST00000391929.7
ENST00000294984.7 ENST00000611909.4 ENST00000367093.3 |
IL24
|
interleukin 24 |
| chrX_+_47078434 | 0.39 |
ENST00000397180.6
|
RGN
|
regucalcin |
| chr1_-_111427731 | 0.38 |
ENST00000369732.4
|
OVGP1
|
oviductal glycoprotein 1 |
| chr16_+_727117 | 0.38 |
ENST00000562141.5
|
HAGHL
|
hydroxyacylglutathione hydrolase like |
| chr13_+_102799104 | 0.37 |
ENST00000639435.1
ENST00000651949.1 ENST00000651008.1 ENST00000257336.6 ENST00000448849.3 |
BIVM-ERCC5
BIVM
|
BIVM-ERCC5 readthrough basic, immunoglobulin-like variable motif containing |
| chr19_+_47337060 | 0.36 |
ENST00000600626.1
|
C5AR2
|
complement component 5a receptor 2 |
| chr17_-_81911350 | 0.36 |
ENST00000570388.5
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr5_-_1886938 | 0.36 |
ENST00000613726.4
|
IRX4
|
iroquois homeobox 4 |
| chr11_+_67289283 | 0.35 |
ENST00000511455.7
|
ANKRD13D
|
ankyrin repeat domain 13D |
| chr11_-_8682619 | 0.35 |
ENST00000646038.2
|
TRIM66
|
tripartite motif containing 66 |
| chr19_-_48673017 | 0.35 |
ENST00000270235.11
ENST00000596844.5 |
NTN5
|
netrin 5 |
| chr19_-_36666810 | 0.34 |
ENST00000592829.5
ENST00000591370.5 ENST00000588268.6 ENST00000360357.8 |
ZNF461
|
zinc finger protein 461 |
| chr2_-_85867641 | 0.34 |
ENST00000393808.8
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr16_-_67483541 | 0.33 |
ENST00000290953.3
|
AGRP
|
agouti related neuropeptide |
| chr16_+_590200 | 0.33 |
ENST00000563109.1
|
RAB40C
|
RAB40C, member RAS oncogene family |
| chr3_+_58237773 | 0.32 |
ENST00000478253.6
|
ABHD6
|
abhydrolase domain containing 6, acylglycerol lipase |
| chr21_-_26845402 | 0.32 |
ENST00000284984.8
ENST00000676955.1 |
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr1_-_11796536 | 0.31 |
ENST00000641820.1
|
MTHFR
|
methylenetetrahydrofolate reductase |
| chr14_-_67488709 | 0.31 |
ENST00000554480.6
|
TMEM229B
|
transmembrane protein 229B |
| chr5_-_67196791 | 0.31 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr19_+_47332167 | 0.30 |
ENST00000595464.3
|
C5AR2
|
complement component 5a receptor 2 |
| chr6_-_41039202 | 0.30 |
ENST00000244565.8
|
UNC5CL
|
unc-5 family C-terminal like |
| chr1_+_32765667 | 0.29 |
ENST00000373480.1
|
KIAA1522
|
KIAA1522 |
| chr12_+_52274610 | 0.29 |
ENST00000423955.7
|
KRT86
|
keratin 86 |
| chr6_+_33410961 | 0.29 |
ENST00000374512.7
ENST00000374516.8 |
PHF1
|
PHD finger protein 1 |
| chr10_+_70478761 | 0.29 |
ENST00000263563.7
|
PALD1
|
phosphatase domain containing paladin 1 |
| chrX_+_154542194 | 0.29 |
ENST00000618670.4
|
IKBKG
|
inhibitor of nuclear factor kappa B kinase regulatory subunit gamma |
| chr1_-_110607816 | 0.28 |
ENST00000485317.6
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr11_-_27700472 | 0.28 |
ENST00000418212.5
ENST00000533246.5 |
BDNF
|
brain derived neurotrophic factor |
| chr2_+_119223815 | 0.28 |
ENST00000393106.6
ENST00000393110.7 ENST00000409811.5 ENST00000393107.2 |
STEAP3
|
STEAP3 metalloreductase |
| chr19_-_48513161 | 0.28 |
ENST00000673139.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr12_+_6494087 | 0.28 |
ENST00000382457.8
ENST00000315579.10 |
NCAPD2
|
non-SMC condensin I complex subunit D2 |
| chr15_+_32717994 | 0.28 |
ENST00000560677.5
ENST00000560830.1 ENST00000651154.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr7_-_122699108 | 0.28 |
ENST00000340112.3
|
RNF133
|
ring finger protein 133 |
| chr15_+_32718476 | 0.27 |
ENST00000652365.1
|
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr1_-_110607970 | 0.27 |
ENST00000638532.1
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr12_+_122774566 | 0.27 |
ENST00000253079.11
|
CCDC62
|
coiled-coil domain containing 62 |
| chr11_+_65639860 | 0.27 |
ENST00000527525.5
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr12_+_122774515 | 0.27 |
ENST00000392441.8
ENST00000539171.1 |
CCDC62
|
coiled-coil domain containing 62 |
| chr19_-_17264718 | 0.27 |
ENST00000431146.6
ENST00000594190.5 |
USHBP1
|
USH1 protein network component harmonin binding protein 1 |
| chr19_-_19170248 | 0.27 |
ENST00000410050.5
ENST00000424583.7 ENST00000409224.5 ENST00000409447.2 |
MEF2B
|
myocyte enhancer factor 2B |
| chr15_-_82709886 | 0.26 |
ENST00000666055.1
ENST00000261722.8 ENST00000535513.2 |
AP3B2
|
adaptor related protein complex 3 subunit beta 2 |
| chr14_-_104953899 | 0.26 |
ENST00000557457.1
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chr16_-_1225257 | 0.26 |
ENST00000234798.4
|
TPSG1
|
tryptase gamma 1 |
| chr22_-_50577915 | 0.25 |
ENST00000360719.6
ENST00000457250.5 |
CPT1B
|
carnitine palmitoyltransferase 1B |
| chr19_-_43082692 | 0.25 |
ENST00000406487.6
|
PSG2
|
pregnancy specific beta-1-glycoprotein 2 |
| chr17_+_44847874 | 0.25 |
ENST00000253410.3
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B |
| chr20_+_45812984 | 0.25 |
ENST00000372568.4
|
UBE2C
|
ubiquitin conjugating enzyme E2 C |
| chr19_+_11355386 | 0.25 |
ENST00000251473.9
ENST00000591329.5 ENST00000586380.5 |
PLPPR2
|
phospholipid phosphatase related 2 |
| chr5_-_150086511 | 0.25 |
ENST00000675795.1
|
CSF1R
|
colony stimulating factor 1 receptor |
| chr19_+_11355491 | 0.25 |
ENST00000591608.1
|
PLPPR2
|
phospholipid phosphatase related 2 |
| chr14_+_100019375 | 0.24 |
ENST00000544450.6
|
EVL
|
Enah/Vasp-like |
| chr11_+_66857056 | 0.24 |
ENST00000309602.5
ENST00000393952.3 |
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr18_-_46657214 | 0.24 |
ENST00000642948.1
|
LOXHD1
|
lipoxygenase homology domains 1 |
| chr3_+_58237501 | 0.24 |
ENST00000295962.8
|
ABHD6
|
abhydrolase domain containing 6, acylglycerol lipase |
| chr16_-_75265925 | 0.24 |
ENST00000535626.6
ENST00000538440.6 ENST00000418647.7 |
BCAR1
|
BCAR1 scaffold protein, Cas family member |
| chr1_+_37793865 | 0.24 |
ENST00000397631.7
|
MANEAL
|
mannosidase endo-alpha like |
| chr1_+_37793787 | 0.24 |
ENST00000373045.11
|
MANEAL
|
mannosidase endo-alpha like |
| chr6_-_41736239 | 0.24 |
ENST00000358871.6
|
TFEB
|
transcription factor EB |
| chr7_+_80133830 | 0.24 |
ENST00000648098.1
ENST00000648476.1 ENST00000648412.1 ENST00000648953.1 ENST00000648306.1 ENST00000648832.1 ENST00000648877.1 ENST00000442586.2 ENST00000649487.1 ENST00000649267.1 |
GNAI1
|
G protein subunit alpha i1 |
| chr8_-_143609547 | 0.24 |
ENST00000433751.5
ENST00000495276.6 ENST00000220966.10 |
PYCR3
|
pyrroline-5-carboxylate reductase 3 |
| chr19_-_35228699 | 0.24 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187 member B |
| chr11_+_3855629 | 0.24 |
ENST00000526596.2
ENST00000300737.8 ENST00000616714.4 |
STIM1
|
stromal interaction molecule 1 |
| chr3_+_50273119 | 0.23 |
ENST00000456560.6
ENST00000418576.3 |
SEMA3B
|
semaphorin 3B |
| chr22_+_22758698 | 0.23 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chrX_-_48071613 | 0.23 |
ENST00000428686.1
ENST00000627643.2 ENST00000276054.9 ENST00000442455.8 |
ZNF630
|
zinc finger protein 630 |
| chr5_-_154850570 | 0.23 |
ENST00000326080.10
ENST00000519501.5 ENST00000518651.5 ENST00000517938.5 ENST00000520461.1 |
FAXDC2
|
fatty acid hydroxylase domain containing 2 |
| chr5_+_175861628 | 0.23 |
ENST00000509837.5
|
CPLX2
|
complexin 2 |
| chr7_-_132576463 | 0.23 |
ENST00000423507.6
|
PLXNA4
|
plexin A4 |
| chr15_-_89751292 | 0.23 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior bHLH transcription factor 1 |
| chr19_+_7497544 | 0.23 |
ENST00000361664.7
|
TEX45
|
testis expressed 45 |
| chr3_+_38282294 | 0.23 |
ENST00000466887.5
ENST00000448498.6 |
SLC22A14
|
solute carrier family 22 member 14 |
| chr12_+_104588845 | 0.23 |
ENST00000549016.1
|
CHST11
|
carbohydrate sulfotransferase 11 |
| chr16_-_48247533 | 0.23 |
ENST00000356608.7
ENST00000569991.1 |
ABCC11
|
ATP binding cassette subfamily C member 11 |
| chr17_+_29593468 | 0.23 |
ENST00000614878.4
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr16_-_28610032 | 0.22 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family 1A member 1 |
| chrX_-_107716401 | 0.22 |
ENST00000486554.1
ENST00000372390.8 |
TSC22D3
|
TSC22 domain family member 3 |
| chr8_+_38030496 | 0.22 |
ENST00000338825.5
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr14_+_24398986 | 0.22 |
ENST00000382554.4
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr17_-_2711633 | 0.22 |
ENST00000435359.5
|
CLUH
|
clustered mitochondria homolog |
| chr22_-_36817001 | 0.22 |
ENST00000406910.6
ENST00000417718.7 |
PVALB
|
parvalbumin |
| chr1_+_966466 | 0.22 |
ENST00000379410.8
ENST00000379407.7 ENST00000379409.6 |
PLEKHN1
|
pleckstrin homology domain containing N1 |
| chr19_-_633500 | 0.22 |
ENST00000588649.7
|
POLRMT
|
RNA polymerase mitochondrial |
| chr16_-_74700786 | 0.22 |
ENST00000306247.11
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain like pseudokinase |
| chr16_+_30199860 | 0.22 |
ENST00000395138.6
|
SULT1A3
|
sulfotransferase family 1A member 3 |
| chr4_-_154590735 | 0.21 |
ENST00000403106.8
ENST00000622532.1 ENST00000651975.1 |
FGA
|
fibrinogen alpha chain |
| chr1_-_205994439 | 0.21 |
ENST00000617991.4
|
RAB7B
|
RAB7B, member RAS oncogene family |
| chr22_+_18110305 | 0.21 |
ENST00000680175.1
ENST00000426208.5 |
TUBA8
|
tubulin alpha 8 |
| chr3_-_149086488 | 0.21 |
ENST00000392912.6
ENST00000465259.5 ENST00000310053.10 ENST00000494055.5 |
HLTF
|
helicase like transcription factor |
| chr19_+_4279247 | 0.21 |
ENST00000543264.7
|
SHD
|
Src homology 2 domain containing transforming protein D |
| chr19_+_10430786 | 0.21 |
ENST00000293683.9
|
PDE4A
|
phosphodiesterase 4A |
| chr19_+_544034 | 0.21 |
ENST00000592501.5
ENST00000264553.6 |
GZMM
|
granzyme M |
| chr12_+_122203681 | 0.21 |
ENST00000546192.1
ENST00000324189.5 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr1_-_16437416 | 0.21 |
ENST00000375577.5
ENST00000335496.5 |
SPATA21
|
spermatogenesis associated 21 |
| chr9_+_137277698 | 0.21 |
ENST00000357503.3
|
TOR4A
|
torsin family 4 member A |
| chr16_+_29459913 | 0.21 |
ENST00000360423.12
|
SULT1A4
|
sulfotransferase family 1A member 4 |
| chr4_-_100190458 | 0.21 |
ENST00000273990.6
|
DDIT4L
|
DNA damage inducible transcript 4 like |
| chr11_+_46332905 | 0.21 |
ENST00000343674.10
|
DGKZ
|
diacylglycerol kinase zeta |
| chr22_-_36817510 | 0.21 |
ENST00000443735.1
|
PVALB
|
parvalbumin |
| chr8_+_144374024 | 0.21 |
ENST00000308860.11
|
ADCK5
|
aarF domain containing kinase 5 |
| chr22_+_30881674 | 0.20 |
ENST00000454145.5
ENST00000453621.5 ENST00000431368.5 ENST00000535268.5 |
OSBP2
|
oxysterol binding protein 2 |
| chr3_-_183555696 | 0.20 |
ENST00000341319.8
|
KLHL6
|
kelch like family member 6 |
| chr1_+_32179665 | 0.20 |
ENST00000373610.8
|
TXLNA
|
taxilin alpha |
| chr1_+_202122881 | 0.20 |
ENST00000683302.1
ENST00000683557.1 |
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr12_-_30754927 | 0.20 |
ENST00000541765.5
ENST00000537108.5 |
CAPRIN2
|
caprin family member 2 |
| chr17_+_80260826 | 0.20 |
ENST00000508628.6
ENST00000582970.5 ENST00000319921.4 |
RNF213
|
ring finger protein 213 |
| chr13_-_98521998 | 0.20 |
ENST00000376547.7
|
STK24
|
serine/threonine kinase 24 |
| chr1_-_209652329 | 0.20 |
ENST00000367030.7
ENST00000356082.9 |
LAMB3
|
laminin subunit beta 3 |
| chr5_-_140346596 | 0.20 |
ENST00000230990.7
|
HBEGF
|
heparin binding EGF like growth factor |
| chr1_-_16437190 | 0.20 |
ENST00000540400.1
|
SPATA21
|
spermatogenesis associated 21 |
| chrX_-_107775951 | 0.20 |
ENST00000315660.8
ENST00000372384.6 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family member 3 |
| chr4_-_15938740 | 0.20 |
ENST00000382333.2
|
FGFBP1
|
fibroblast growth factor binding protein 1 |
| chr11_+_94543894 | 0.19 |
ENST00000358752.4
|
FUT4
|
fucosyltransferase 4 |
| chrX_-_70260199 | 0.19 |
ENST00000374519.4
|
P2RY4
|
pyrimidinergic receptor P2Y4 |
| chr2_-_96505345 | 0.19 |
ENST00000310865.7
ENST00000451794.6 |
NEURL3
|
neuralized E3 ubiquitin protein ligase 3 |
| chr19_-_17264732 | 0.19 |
ENST00000252597.8
|
USHBP1
|
USH1 protein network component harmonin binding protein 1 |
| chr16_+_30199255 | 0.19 |
ENST00000338971.10
|
SULT1A3
|
sulfotransferase family 1A member 3 |
| chr1_+_32180044 | 0.19 |
ENST00000373609.1
|
TXLNA
|
taxilin alpha |
| chr17_-_74868616 | 0.19 |
ENST00000579893.1
ENST00000544854.5 |
FDXR
|
ferredoxin reductase |
| chr17_-_81911200 | 0.19 |
ENST00000570391.5
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr5_+_32710630 | 0.19 |
ENST00000326958.5
|
NPR3
|
natriuretic peptide receptor 3 |
| chr6_+_31615215 | 0.19 |
ENST00000337917.11
ENST00000376059.8 |
AIF1
|
allograft inflammatory factor 1 |
| chr4_-_80073057 | 0.19 |
ENST00000681710.1
|
ANTXR2
|
ANTXR cell adhesion molecule 2 |
| chr22_+_30694851 | 0.19 |
ENST00000332585.11
|
OSBP2
|
oxysterol binding protein 2 |
| chr7_+_143381561 | 0.19 |
ENST00000354434.8
|
ZYX
|
zyxin |
| chr15_-_73753308 | 0.19 |
ENST00000569673.3
|
INSYN1
|
inhibitory synaptic factor 1 |
| chr17_-_74531467 | 0.19 |
ENST00000314401.3
ENST00000392621.6 |
CD300LB
|
CD300 molecule like family member b |
| chr7_-_27152561 | 0.19 |
ENST00000467897.2
ENST00000612286.5 ENST00000498652.1 |
ENSG00000273433.1
HOXA3
ENSG00000272801.1
|
novel transcript homeobox A3 novel transcript |
| chr14_+_94110728 | 0.18 |
ENST00000616764.4
ENST00000618863.1 ENST00000611954.4 ENST00000618200.4 ENST00000621160.4 ENST00000555819.5 ENST00000620396.4 ENST00000612813.4 ENST00000620066.1 |
IFI27
|
interferon alpha inducible protein 27 |
| chr22_-_38755458 | 0.18 |
ENST00000405510.5
ENST00000433561.5 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr16_-_74700845 | 0.18 |
ENST00000308807.12
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain like pseudokinase |
| chr7_-_16421524 | 0.18 |
ENST00000407010.7
|
CRPPA
|
CDP-L-ribitol pyrophosphorylase A |
| chr16_-_1230089 | 0.18 |
ENST00000612142.1
ENST00000606293.5 |
TPSB2
|
tryptase beta 2 |
| chr14_+_89955895 | 0.18 |
ENST00000393452.7
ENST00000554180.5 ENST00000335725.9 ENST00000393454.6 ENST00000553617.5 ENST00000556867.1 ENST00000553527.1 |
TDP1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr3_+_129548386 | 0.18 |
ENST00000503977.1
|
H1-8
|
H1.8 linker histone |
| chr8_+_73294594 | 0.18 |
ENST00000240285.10
|
RDH10
|
retinol dehydrogenase 10 |
| chr7_+_143381907 | 0.18 |
ENST00000392910.6
|
ZYX
|
zyxin |
| chr19_+_41350911 | 0.18 |
ENST00000539627.5
|
TMEM91
|
transmembrane protein 91 |
| chrX_-_153971169 | 0.18 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 |
| chr1_-_53738024 | 0.17 |
ENST00000628545.1
|
GLIS1
|
GLIS family zinc finger 1 |
| chr3_+_39383337 | 0.17 |
ENST00000650617.1
ENST00000431510.1 ENST00000645630.1 |
SLC25A38
|
solute carrier family 25 member 38 |
| chr3_+_48446710 | 0.17 |
ENST00000635099.1
ENST00000346691.9 ENST00000320211.10 |
ATRIP
|
ATR interacting protein |
| chr10_-_107164692 | 0.17 |
ENST00000263054.11
|
SORCS1
|
sortilin related VPS10 domain containing receptor 1 |
| chr1_+_42825548 | 0.17 |
ENST00000372514.7
|
ERMAP
|
erythroblast membrane associated protein (Scianna blood group) |
| chr19_-_15331890 | 0.17 |
ENST00000594841.5
ENST00000601941.1 |
BRD4
|
bromodomain containing 4 |
| chr7_-_150341615 | 0.17 |
ENST00000223271.8
ENST00000466675.5 ENST00000482669.1 ENST00000467793.5 |
RARRES2
|
retinoic acid receptor responder 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.3 | 1.2 | GO:1903629 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 0.3 | 0.8 | GO:0014028 | notochord formation(GO:0014028) |
| 0.2 | 0.7 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.2 | 0.7 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.2 | 0.7 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.2 | 0.6 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.2 | 2.4 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 0.5 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 0.4 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.6 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 1.6 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.8 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.4 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.1 | 0.5 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.2 | GO:0042662 | negative regulation of mesodermal cell fate specification(GO:0042662) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.4 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.3 | GO:1902823 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.2 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.1 | 0.2 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.1 | 0.3 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.2 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.1 | 0.4 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.3 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.1 | 0.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.2 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 0.6 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.2 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.3 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.2 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.0 | 0.4 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 5.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.5 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.3 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.2 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.0 | 0.1 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 1.1 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.1 | GO:0043016 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.0 | 0.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.4 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.0 | 0.4 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.2 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.3 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.2 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0001794 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 0.0 | 0.1 | GO:0051586 | peptidyl-cysteine methylation(GO:0018125) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.2 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) sodium-dependent phosphate transport(GO:0044341) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.2 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.4 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.1 | GO:0044010 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.1 | GO:1902563 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.0 | 0.1 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.2 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.0 | 0.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.2 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.0 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.0 | GO:0018963 | phthalate metabolic process(GO:0018963) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.5 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.0 | 0.0 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.3 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.1 | 0.4 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 0.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.3 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 1.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.4 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 2.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.2 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.0 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.0 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.3 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.2 | 0.7 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.2 | 2.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.7 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 0.8 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.2 | 0.6 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 1.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.4 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.5 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.6 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.3 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 0.3 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 0.2 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 0.2 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 0.1 | 0.3 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.4 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 0.2 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.2 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.0 | 0.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.5 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.4 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.2 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0047693 | GTP diphosphatase activity(GO:0036219) 2-hydroxy-adenosine triphosphate pyrophosphatase activity(GO:0044713) 2-hydroxy-(deoxy)adenosine-triphosphate pyrophosphatase activity(GO:0044714) ATP diphosphatase activity(GO:0047693) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 5.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.3 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0032396 | MHC class Ib protein binding(GO:0023029) inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.0 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 3.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.8 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |