Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for EZH2
Z-value: 0.73
Transcription factors associated with EZH2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EZH2
|
ENSG00000106462.12 | EZH2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EZH2 | hg38_v1_chr7_-_148883474_148883509 | 0.18 | 3.9e-01 | Click! |
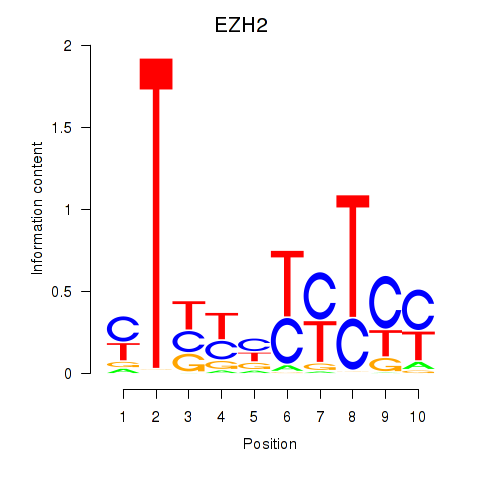
Activity profile of EZH2 motif
Sorted Z-values of EZH2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of EZH2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_31582415 | 1.87 |
ENST00000429299.3
ENST00000446745.2 |
LTB
|
lymphotoxin beta |
| chr19_+_50649445 | 1.02 |
ENST00000425202.6
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr11_-_44310126 | 0.99 |
ENST00000652299.1
|
ALX4
|
ALX homeobox 4 |
| chr11_-_8810635 | 0.96 |
ENST00000527510.5
ENST00000528527.5 ENST00000313726.11 ENST00000528523.5 |
DENND2B
|
DENN domain containing 2B |
| chr9_-_16728165 | 0.95 |
ENST00000603713.5
ENST00000603313.5 |
BNC2
|
basonuclin 2 |
| chr4_-_76023489 | 0.81 |
ENST00000306602.3
|
CXCL10
|
C-X-C motif chemokine ligand 10 |
| chr19_+_45001430 | 0.81 |
ENST00000625761.2
ENST00000505236.1 ENST00000221452.13 |
RELB
|
RELB proto-oncogene, NF-kB subunit |
| chr14_+_23376765 | 0.77 |
ENST00000649278.1
|
CMTM5
|
CKLF like MARVEL transmembrane domain containing 5 |
| chr14_-_75981986 | 0.65 |
ENST00000238682.8
|
TGFB3
|
transforming growth factor beta 3 |
| chr7_-_22193728 | 0.63 |
ENST00000620335.4
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr2_+_227813834 | 0.59 |
ENST00000358813.5
ENST00000409189.7 |
CCL20
|
C-C motif chemokine ligand 20 |
| chr6_-_10412367 | 0.58 |
ENST00000379608.9
|
TFAP2A
|
transcription factor AP-2 alpha |
| chr5_-_147081428 | 0.58 |
ENST00000394413.7
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr8_+_32647080 | 0.56 |
ENST00000520502.7
ENST00000523041.2 ENST00000650819.1 |
NRG1
|
neuregulin 1 |
| chr7_-_22193824 | 0.56 |
ENST00000401957.6
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr1_+_209686173 | 0.55 |
ENST00000615289.4
ENST00000367028.6 ENST00000261465.5 |
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr14_+_23376786 | 0.54 |
ENST00000359320.7
|
CMTM5
|
CKLF like MARVEL transmembrane domain containing 5 |
| chr17_+_7435416 | 0.53 |
ENST00000323206.2
ENST00000396568.1 |
TMEM102
|
transmembrane protein 102 |
| chr18_+_74499939 | 0.51 |
ENST00000584768.5
|
CNDP2
|
carnosine dipeptidase 2 |
| chr6_+_36676455 | 0.51 |
ENST00000615513.4
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr3_+_122680802 | 0.50 |
ENST00000474629.7
|
PARP14
|
poly(ADP-ribose) polymerase family member 14 |
| chr20_-_675793 | 0.50 |
ENST00000488788.2
ENST00000246104.7 |
ENSG00000270299.1
SCRT2
|
novel protein scratch family transcriptional repressor 2 |
| chr14_+_75605462 | 0.49 |
ENST00000539311.5
|
FLVCR2
|
FLVCR heme transporter 2 |
| chr14_+_23377001 | 0.48 |
ENST00000342473.8
ENST00000397227.7 ENST00000339180.9 ENST00000555731.5 |
CMTM5
|
CKLF like MARVEL transmembrane domain containing 5 |
| chr11_+_6205549 | 0.47 |
ENST00000316375.3
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr2_+_96537254 | 0.46 |
ENST00000454558.2
|
ARID5A
|
AT-rich interaction domain 5A |
| chr9_+_114155526 | 0.45 |
ENST00000356083.8
|
COL27A1
|
collagen type XXVII alpha 1 chain |
| chr3_-_158106050 | 0.44 |
ENST00000441443.6
ENST00000389589.8 |
SHOX2
|
short stature homeobox 2 |
| chr5_-_147081462 | 0.44 |
ENST00000508267.5
ENST00000504198.5 |
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr14_+_21070273 | 0.43 |
ENST00000555038.5
ENST00000298694.9 |
ARHGEF40
|
Rho guanine nucleotide exchange factor 40 |
| chr3_-_108058361 | 0.43 |
ENST00000398258.7
|
CD47
|
CD47 molecule |
| chr1_+_13585453 | 0.43 |
ENST00000487038.5
ENST00000475043.5 |
PDPN
|
podoplanin |
| chr4_-_145938473 | 0.42 |
ENST00000513320.5
|
ZNF827
|
zinc finger protein 827 |
| chr6_+_36676489 | 0.42 |
ENST00000448526.6
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr6_+_137871208 | 0.41 |
ENST00000614035.4
ENST00000621150.3 ENST00000619035.4 ENST00000615468.4 ENST00000620204.3 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr14_+_103123452 | 0.41 |
ENST00000558056.1
ENST00000560869.6 |
TNFAIP2
|
TNF alpha induced protein 2 |
| chr12_+_6828377 | 0.41 |
ENST00000290510.10
|
P3H3
|
prolyl 3-hydroxylase 3 |
| chr22_+_22030934 | 0.40 |
ENST00000390282.2
|
IGLV4-69
|
immunoglobulin lambda variable 4-69 |
| chr12_+_26195647 | 0.40 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr1_-_56579555 | 0.39 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr12_+_10179024 | 0.39 |
ENST00000543484.2
|
TMEM52B
|
transmembrane protein 52B |
| chr2_-_144520106 | 0.38 |
ENST00000627532.3
ENST00000539609.7 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr3_-_158106408 | 0.38 |
ENST00000483851.7
|
SHOX2
|
short stature homeobox 2 |
| chr6_-_30686624 | 0.37 |
ENST00000274853.8
|
PPP1R18
|
protein phosphatase 1 regulatory subunit 18 |
| chr11_+_64237420 | 0.37 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr5_+_175871670 | 0.37 |
ENST00000514150.5
|
CPLX2
|
complexin 2 |
| chr12_+_10179006 | 0.34 |
ENST00000298530.7
|
TMEM52B
|
transmembrane protein 52B |
| chr16_-_84618067 | 0.34 |
ENST00000262428.5
|
COTL1
|
coactosin like F-actin binding protein 1 |
| chr5_-_150289941 | 0.34 |
ENST00000682786.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr7_+_144003929 | 0.34 |
ENST00000408922.3
|
OR6B1
|
olfactory receptor family 6 subfamily B member 1 |
| chr1_-_173207322 | 0.34 |
ENST00000281834.4
|
TNFSF4
|
TNF superfamily member 4 |
| chr6_+_125919296 | 0.34 |
ENST00000444128.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr6_+_125919210 | 0.33 |
ENST00000438495.6
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr19_+_53909263 | 0.33 |
ENST00000391767.6
|
CACNG7
|
calcium voltage-gated channel auxiliary subunit gamma 7 |
| chr16_-_84618041 | 0.33 |
ENST00000564057.1
|
COTL1
|
coactosin like F-actin binding protein 1 |
| chr2_+_63050813 | 0.33 |
ENST00000282549.7
|
OTX1
|
orthodenticle homeobox 1 |
| chr9_-_131709721 | 0.33 |
ENST00000372190.7
|
RAPGEF1
|
Rap guanine nucleotide exchange factor 1 |
| chr11_-_107712049 | 0.32 |
ENST00000305991.3
|
SLN
|
sarcolipin |
| chr5_-_160685379 | 0.31 |
ENST00000642502.1
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr8_-_167024 | 0.31 |
ENST00000320901.4
|
OR4F21
|
olfactory receptor family 4 subfamily F member 21 |
| chr2_+_90069662 | 0.31 |
ENST00000390271.2
|
IGKV6D-41
|
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr9_+_122510802 | 0.30 |
ENST00000335302.5
|
OR1J2
|
olfactory receptor family 1 subfamily J member 2 |
| chr18_-_3219961 | 0.30 |
ENST00000356443.9
|
MYOM1
|
myomesin 1 |
| chr12_-_75209701 | 0.30 |
ENST00000350228.6
ENST00000298972.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr12_-_75209814 | 0.29 |
ENST00000549446.6
|
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr5_-_35938572 | 0.29 |
ENST00000651391.1
ENST00000397366.5 ENST00000513623.5 ENST00000514524.2 ENST00000397367.6 |
CAPSL
|
calcyphosine like |
| chr8_+_125430333 | 0.29 |
ENST00000311922.4
|
TRIB1
|
tribbles pseudokinase 1 |
| chr2_+_90021567 | 0.28 |
ENST00000436451.2
|
IGKV6D-21
|
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr11_-_72674394 | 0.28 |
ENST00000418754.6
ENST00000334456.10 ENST00000542969.2 |
PDE2A
|
phosphodiesterase 2A |
| chr17_+_7484357 | 0.28 |
ENST00000674977.2
|
POLR2A
|
RNA polymerase II subunit A |
| chr11_-_34511710 | 0.28 |
ENST00000620316.4
ENST00000312319.6 |
ELF5
|
E74 like ETS transcription factor 5 |
| chr17_+_48723179 | 0.28 |
ENST00000422730.4
|
PRAC2
|
PRAC2 small nuclear protein |
| chr16_-_67247460 | 0.28 |
ENST00000258201.9
|
FHOD1
|
formin homology 2 domain containing 1 |
| chr5_+_175871570 | 0.28 |
ENST00000512824.5
ENST00000393745.8 |
CPLX2
|
complexin 2 |
| chr15_+_84237586 | 0.28 |
ENST00000512109.1
|
GOLGA6L4
|
golgin A6 family like 4 |
| chr21_+_25639251 | 0.28 |
ENST00000480456.6
|
JAM2
|
junctional adhesion molecule 2 |
| chr17_+_7650916 | 0.27 |
ENST00000250111.9
|
ATP1B2
|
ATPase Na+/K+ transporting subunit beta 2 |
| chr1_-_209651291 | 0.27 |
ENST00000391911.5
ENST00000415782.1 |
LAMB3
|
laminin subunit beta 3 |
| chr9_-_16727980 | 0.27 |
ENST00000418777.5
ENST00000468187.6 |
BNC2
|
basonuclin 2 |
| chr1_+_175067831 | 0.27 |
ENST00000239462.9
|
TNN
|
tenascin N |
| chr5_-_150289625 | 0.27 |
ENST00000683332.1
ENST00000398376.8 ENST00000672785.1 ENST00000672396.1 |
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr12_+_6821797 | 0.27 |
ENST00000311268.8
ENST00000382315.7 |
GPR162
|
G protein-coupled receptor 162 |
| chr1_-_153549238 | 0.27 |
ENST00000368713.8
|
S100A3
|
S100 calcium binding protein A3 |
| chr1_-_120100688 | 0.26 |
ENST00000652264.1
|
NOTCH2
|
notch receptor 2 |
| chrX_-_25015924 | 0.26 |
ENST00000379044.5
|
ARX
|
aristaless related homeobox |
| chr19_-_47545261 | 0.26 |
ENST00000595558.1
ENST00000263351.9 |
ZNF541
|
zinc finger protein 541 |
| chr6_-_42722590 | 0.26 |
ENST00000230381.7
|
PRPH2
|
peripherin 2 |
| chr4_-_145938422 | 0.26 |
ENST00000656985.1
ENST00000652097.1 ENST00000503462.3 ENST00000379448.9 ENST00000513840.2 |
ZNF827
|
zinc finger protein 827 |
| chr15_+_81000913 | 0.26 |
ENST00000267984.4
|
TLNRD1
|
talin rod domain containing 1 |
| chr2_+_165469647 | 0.25 |
ENST00000421875.5
ENST00000314499.11 ENST00000409664.5 ENST00000651982.1 |
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr19_+_38647679 | 0.25 |
ENST00000390009.7
ENST00000589528.1 |
ACTN4
|
actinin alpha 4 |
| chr2_+_201129483 | 0.25 |
ENST00000440180.5
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr1_+_156154371 | 0.24 |
ENST00000368282.1
|
SEMA4A
|
semaphorin 4A |
| chr11_-_74731148 | 0.24 |
ENST00000263671.9
ENST00000528789.1 |
CHRDL2
|
chordin like 2 |
| chr21_+_25639272 | 0.24 |
ENST00000400532.5
ENST00000312957.9 |
JAM2
|
junctional adhesion molecule 2 |
| chr18_-_3219849 | 0.24 |
ENST00000261606.11
|
MYOM1
|
myomesin 1 |
| chr16_+_31074390 | 0.24 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr1_+_100538131 | 0.24 |
ENST00000315033.5
|
GPR88
|
G protein-coupled receptor 88 |
| chr10_+_97589715 | 0.24 |
ENST00000370640.5
|
C10orf62
|
chromosome 10 open reading frame 62 |
| chr3_+_100609594 | 0.24 |
ENST00000273352.8
|
ADGRG7
|
adhesion G protein-coupled receptor G7 |
| chr14_+_24094053 | 0.24 |
ENST00000559250.5
ENST00000560736.5 ENST00000396973.8 ENST00000559837.5 |
PCK2
|
phosphoenolpyruvate carboxykinase 2, mitochondrial |
| chr1_-_153549120 | 0.24 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr2_+_201129318 | 0.23 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr4_-_167234266 | 0.23 |
ENST00000511269.5
ENST00000506697.5 ENST00000512042.1 |
SPOCK3
|
SPARC (osteonectin), cwcv and kazal like domains proteoglycan 3 |
| chr1_-_158426237 | 0.23 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr1_+_154220849 | 0.23 |
ENST00000613315.4
ENST00000271877.11 ENST00000441890.5 ENST00000412596.5 ENST00000368504.5 ENST00000428931.6 ENST00000437652.5 |
UBAP2L
|
ubiquitin associated protein 2 like |
| chr5_+_153491174 | 0.23 |
ENST00000521843.6
|
GRIA1
|
glutamate ionotropic receptor AMPA type subunit 1 |
| chr17_+_34319427 | 0.23 |
ENST00000394620.2
|
CCL8
|
C-C motif chemokine ligand 8 |
| chr16_-_11527245 | 0.23 |
ENST00000598234.6
|
ENSG00000188897.9
|
novel lipoprotein amino terminal region containing protein |
| chr4_+_56505782 | 0.23 |
ENST00000640821.3
|
ARL9
|
ADP ribosylation factor like GTPase 9 |
| chr11_-_35419462 | 0.23 |
ENST00000643522.1
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr2_+_90114838 | 0.23 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 |
| chr3_-_195811889 | 0.23 |
ENST00000475231.5
|
MUC4
|
mucin 4, cell surface associated |
| chr11_-_35419213 | 0.23 |
ENST00000642171.1
ENST00000644050.1 ENST00000643134.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr11_-_35418966 | 0.23 |
ENST00000531628.2
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr17_+_36103819 | 0.23 |
ENST00000615863.2
ENST00000621626.1 |
CCL4
|
C-C motif chemokine ligand 4 |
| chr19_+_38647614 | 0.22 |
ENST00000252699.7
ENST00000424234.7 ENST00000440400.2 |
ACTN4
|
actinin alpha 4 |
| chrX_-_132489842 | 0.22 |
ENST00000436215.5
|
MBNL3
|
muscleblind like splicing regulator 3 |
| chr6_+_108166015 | 0.22 |
ENST00000368986.9
|
NR2E1
|
nuclear receptor subfamily 2 group E member 1 |
| chr3_+_19148500 | 0.22 |
ENST00000328405.7
|
KCNH8
|
potassium voltage-gated channel subfamily H member 8 |
| chr5_-_143403297 | 0.22 |
ENST00000415690.6
|
NR3C1
|
nuclear receptor subfamily 3 group C member 1 |
| chr16_+_50742037 | 0.22 |
ENST00000569418.5
|
CYLD
|
CYLD lysine 63 deubiquitinase |
| chr16_-_89719369 | 0.22 |
ENST00000561976.5
|
VPS9D1
|
VPS9 domain containing 1 |
| chr3_+_111542134 | 0.22 |
ENST00000438817.6
|
CD96
|
CD96 molecule |
| chr3_-_129316276 | 0.22 |
ENST00000333762.6
|
H1-10
|
H1.10 linker histone |
| chr4_-_48080172 | 0.22 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr5_-_136365476 | 0.22 |
ENST00000378459.7
ENST00000502753.4 ENST00000513104.6 ENST00000352189.8 |
TRPC7
|
transient receptor potential cation channel subfamily C member 7 |
| chr5_+_35856883 | 0.21 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr6_-_32178080 | 0.21 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr19_+_42076129 | 0.21 |
ENST00000359044.5
|
ZNF574
|
zinc finger protein 574 |
| chr8_-_100309368 | 0.21 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr5_-_177509843 | 0.21 |
ENST00000510380.5
ENST00000357198.9 |
DOK3
|
docking protein 3 |
| chr15_+_59611776 | 0.21 |
ENST00000396065.3
ENST00000560585.5 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr1_+_203475798 | 0.21 |
ENST00000343110.3
|
PRELP
|
proline and arginine rich end leucine rich repeat protein |
| chr12_+_6821646 | 0.21 |
ENST00000428545.6
|
GPR162
|
G protein-coupled receptor 162 |
| chr17_+_4833331 | 0.21 |
ENST00000355280.11
ENST00000347992.11 |
MINK1
|
misshapen like kinase 1 |
| chr3_-_171026709 | 0.20 |
ENST00000314251.8
|
SLC2A2
|
solute carrier family 2 member 2 |
| chr5_-_147782518 | 0.20 |
ENST00000507386.5
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr1_+_147902789 | 0.20 |
ENST00000369235.2
|
GJA8
|
gap junction protein alpha 8 |
| chr3_-_71130892 | 0.20 |
ENST00000491238.7
ENST00000674446.1 |
FOXP1
|
forkhead box P1 |
| chr11_-_35419098 | 0.20 |
ENST00000606205.6
ENST00000645303.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr16_+_7304219 | 0.20 |
ENST00000675562.1
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr8_-_144336451 | 0.20 |
ENST00000569446.3
|
SCRT1
|
scratch family transcriptional repressor 1 |
| chr2_-_144520054 | 0.20 |
ENST00000419938.5
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_-_150289764 | 0.20 |
ENST00000671881.1
ENST00000672752.1 ENST00000510347.2 ENST00000672829.1 ENST00000348628.11 |
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha |
| chr7_-_155812454 | 0.20 |
ENST00000297261.7
|
SHH
|
sonic hedgehog signaling molecule |
| chr16_-_29923237 | 0.20 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr2_+_90172802 | 0.20 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr17_+_38705482 | 0.20 |
ENST00000620609.4
|
MLLT6
|
MLLT6, PHD finger containing |
| chr4_+_158521937 | 0.20 |
ENST00000343542.9
ENST00000470033.2 |
RXFP1
|
relaxin family peptide receptor 1 |
| chr17_+_34255274 | 0.20 |
ENST00000580907.5
ENST00000225831.4 |
CCL2
|
C-C motif chemokine ligand 2 |
| chr3_-_71130963 | 0.20 |
ENST00000649695.2
|
FOXP1
|
forkhead box P1 |
| chr16_+_30650932 | 0.20 |
ENST00000300835.9
|
PRR14
|
proline rich 14 |
| chr17_-_75848641 | 0.20 |
ENST00000586257.5
|
WBP2
|
WW domain binding protein 2 |
| chr7_+_20330893 | 0.20 |
ENST00000222573.5
|
ITGB8
|
integrin subunit beta 8 |
| chr2_+_96536743 | 0.20 |
ENST00000673792.1
ENST00000357485.8 |
ARID5A
|
AT-rich interaction domain 5A |
| chr15_-_77696142 | 0.19 |
ENST00000561030.5
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr12_-_69699353 | 0.19 |
ENST00000331471.8
|
BEST3
|
bestrophin 3 |
| chr12_-_69699331 | 0.19 |
ENST00000548658.1
ENST00000476098.5 |
BEST3
|
bestrophin 3 |
| chr1_+_165827574 | 0.19 |
ENST00000367879.9
|
UCK2
|
uridine-cytidine kinase 2 |
| chr10_+_73911104 | 0.19 |
ENST00000446342.5
ENST00000372764.4 |
PLAU
|
plasminogen activator, urokinase |
| chr16_+_30651039 | 0.19 |
ENST00000542965.2
|
PRR14
|
proline rich 14 |
| chrX_-_48958348 | 0.19 |
ENST00000428668.2
|
OTUD5
|
OTU deubiquitinase 5 |
| chr3_+_133400046 | 0.19 |
ENST00000302334.3
|
BFSP2
|
beaded filament structural protein 2 |
| chr10_+_19489054 | 0.19 |
ENST00000377265.3
|
MALRD1
|
MAM and LDL receptor class A domain containing 1 |
| chr4_+_24795560 | 0.19 |
ENST00000382120.4
|
SOD3
|
superoxide dismutase 3 |
| chr2_-_222320124 | 0.18 |
ENST00000678139.1
|
ENSG00000288658.1
|
novel protein, ortholog of Gm2102 (M. musculus) |
| chr9_+_116153783 | 0.18 |
ENST00000328252.4
|
PAPPA
|
pappalysin 1 |
| chr4_-_47981535 | 0.18 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr15_-_37098281 | 0.18 |
ENST00000559085.5
ENST00000397624.7 |
MEIS2
|
Meis homeobox 2 |
| chr12_+_20695323 | 0.18 |
ENST00000266509.7
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr6_-_165986506 | 0.18 |
ENST00000676766.1
ENST00000647768.3 |
PDE10A
|
phosphodiesterase 10A |
| chr17_-_62808339 | 0.18 |
ENST00000583600.5
|
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chr17_-_58328756 | 0.18 |
ENST00000268893.10
ENST00000343736.9 |
TSPOAP1
|
TSPO associated protein 1 |
| chr17_-_62808299 | 0.18 |
ENST00000311269.10
|
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chr17_-_37745018 | 0.18 |
ENST00000613727.4
ENST00000614313.4 ENST00000617811.5 ENST00000621123.4 |
HNF1B
|
HNF1 homeobox B |
| chr22_-_45212431 | 0.18 |
ENST00000496226.1
ENST00000251993.11 |
KIAA0930
|
KIAA0930 |
| chr4_+_73404255 | 0.17 |
ENST00000621628.4
ENST00000621085.4 ENST00000415165.6 ENST00000295897.9 ENST00000503124.5 ENST00000509063.5 ENST00000401494.7 |
ALB
|
albumin |
| chr2_-_127107144 | 0.17 |
ENST00000316724.10
|
BIN1
|
bridging integrator 1 |
| chr10_-_73641450 | 0.17 |
ENST00000359322.5
|
MYOZ1
|
myozenin 1 |
| chr19_+_56595279 | 0.17 |
ENST00000328070.10
ENST00000599599.7 ENST00000650950.1 |
ZNF71
ZIM2-AS1
|
zinc finger protein 71 ZIM2 antisense RNA 1 |
| chr17_-_38748184 | 0.17 |
ENST00000618941.4
ENST00000620225.5 ENST00000618506.1 ENST00000616129.4 |
PCGF2
|
polycomb group ring finger 2 |
| chr3_-_15521675 | 0.17 |
ENST00000383788.10
ENST00000383786.9 ENST00000603808.5 |
COLQ
|
collagen like tail subunit of asymmetric acetylcholinesterase |
| chr3_+_101849505 | 0.17 |
ENST00000326151.9
ENST00000326172.9 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr5_-_77638713 | 0.17 |
ENST00000306422.5
|
OTP
|
orthopedia homeobox |
| chr12_+_56230043 | 0.17 |
ENST00000424625.5
ENST00000419753.5 ENST00000454355.7 ENST00000417965.5 ENST00000436633.5 |
SLC39A5
|
solute carrier family 39 member 5 |
| chr17_-_2336435 | 0.17 |
ENST00000301364.10
ENST00000576112.2 |
TSR1
|
TSR1 ribosome maturation factor |
| chr19_-_39391029 | 0.17 |
ENST00000221265.8
|
PAF1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr2_-_144516397 | 0.17 |
ENST00000638128.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr11_+_66421004 | 0.17 |
ENST00000639555.1
ENST00000311034.7 |
NPAS4
|
neuronal PAS domain protein 4 |
| chr15_+_42359577 | 0.17 |
ENST00000357568.8
|
CAPN3
|
calpain 3 |
| chr14_+_24094301 | 0.17 |
ENST00000545054.6
ENST00000561286.5 ENST00000216780.9 ENST00000558096.5 |
PCK2
|
phosphoenolpyruvate carboxykinase 2, mitochondrial |
| chr2_+_1414382 | 0.16 |
ENST00000423320.5
ENST00000346956.7 ENST00000382198.5 |
TPO
|
thyroid peroxidase |
| chr8_+_32646838 | 0.16 |
ENST00000651333.1
ENST00000652592.1 |
NRG1
|
neuregulin 1 |
| chr10_-_59362584 | 0.16 |
ENST00000618427.4
ENST00000611933.4 |
FAM13C
|
family with sequence similarity 13 member C |
| chr17_+_38705243 | 0.16 |
ENST00000621332.5
|
MLLT6
|
MLLT6, PHD finger containing |
| chr14_+_22836574 | 0.16 |
ENST00000548162.2
ENST00000311852.11 |
MMP14
|
matrix metallopeptidase 14 |
| chr10_+_73744346 | 0.16 |
ENST00000345254.9
ENST00000339365.2 |
SEC24C
|
SEC24 homolog C, COPII coat complex component |
| chr17_+_34285734 | 0.16 |
ENST00000305869.4
|
CCL11
|
C-C motif chemokine ligand 11 |
| chr6_-_29633171 | 0.16 |
ENST00000377034.9
|
GABBR1
|
gamma-aminobutyric acid type B receptor subunit 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.2 | 1.9 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 0.6 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 0.7 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.2 | 0.5 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.4 | GO:0034147 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.1 | 0.4 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 1.4 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.3 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.3 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.9 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.3 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.1 | GO:2001303 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.1 | 0.3 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.2 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) |
| 0.1 | 0.3 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.5 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.3 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.1 | 0.4 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.3 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.1 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.2 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.2 | GO:1990451 | cellular stress response to acidic pH(GO:1990451) |
| 0.1 | 0.3 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.1 | 0.9 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.2 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.1 | 0.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.2 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 1.0 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.3 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.7 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.2 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.1 | 0.6 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 0.3 | GO:2000501 | regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.5 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 2.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.3 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.2 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0003166 | bundle of His development(GO:0003166) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.4 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:0043006 | reduction of food intake in response to dietary excess(GO:0002023) activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.3 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.8 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.1 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 1.3 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.2 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.2 | GO:0060529 | ectoderm and mesoderm interaction(GO:0007499) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.8 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.3 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:1903216 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.0 | 0.2 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.0 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.0 | 0.2 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0071931 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.0 | 0.1 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.8 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.6 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.8 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.1 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.0 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.1 | GO:0048687 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.2 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.0 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.1 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.2 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.0 | GO:0002588 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.1 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.0 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0001810 | regulation of type I hypersensitivity(GO:0001810) positive regulation of type I hypersensitivity(GO:0001812) B cell selection(GO:0002339) B cell cytokine production(GO:0002368) type I hypersensitivity(GO:0016068) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.0 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.0 | 0.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.1 | GO:0060355 | regulation of cell adhesion molecule production(GO:0060353) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.4 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.0 | GO:0100009 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.0 | 0.2 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.0 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.6 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.9 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.0 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.2 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.2 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.0 | GO:2000118 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.0 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0061205 | alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.0 | 0.0 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.0 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.0 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0086073 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.2 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.5 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.3 | GO:0060987 | lipid tube(GO:0060987) |
| 0.1 | 0.6 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.2 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.8 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 2.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.6 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.0 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 1.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.2 | 0.6 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 0.9 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 0.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 0.5 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 0.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 1.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.3 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 1.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.2 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 0.5 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.9 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 1.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.5 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.8 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.6 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.0 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 5.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 2.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |