Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for FIGLA
Z-value: 0.76
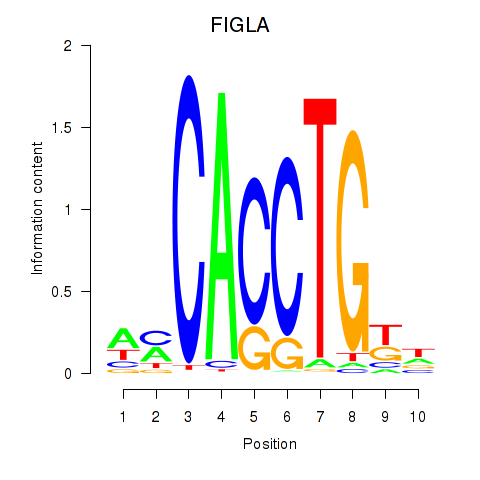
Transcription factors associated with FIGLA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FIGLA
|
ENSG00000183733.6 | FIGLA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FIGLA | hg38_v1_chr2_-_70790643_70790643 | 0.07 | 7.3e-01 | Click! |
Activity profile of FIGLA motif
Sorted Z-values of FIGLA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of FIGLA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_41226648 | 5.28 |
ENST00000377721.3
|
KRTAP9-2
|
keratin associated protein 9-2 |
| chr17_+_41237998 | 4.87 |
ENST00000254072.7
|
KRTAP9-8
|
keratin associated protein 9-8 |
| chr6_-_31582415 | 2.13 |
ENST00000429299.3
ENST00000446745.2 |
LTB
|
lymphotoxin beta |
| chr1_-_12616762 | 1.95 |
ENST00000464917.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr16_-_46831043 | 1.39 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr17_+_41105332 | 1.24 |
ENST00000391415.1
ENST00000617453.1 |
KRTAP4-9
|
keratin associated protein 4-9 |
| chr7_-_11832190 | 1.00 |
ENST00000423059.9
ENST00000617773.1 |
THSD7A
|
thrombospondin type 1 domain containing 7A |
| chr22_-_42720813 | 0.91 |
ENST00000381278.4
|
A4GALT
|
alpha 1,4-galactosyltransferase (P blood group) |
| chr22_-_42720861 | 0.90 |
ENST00000642412.2
|
A4GALT
|
alpha 1,4-galactosyltransferase (P blood group) |
| chr14_-_54902807 | 0.86 |
ENST00000543643.6
ENST00000536224.2 ENST00000395514.5 ENST00000491895.7 |
GCH1
|
GTP cyclohydrolase 1 |
| chr6_-_31728877 | 0.78 |
ENST00000437288.5
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr17_+_70169516 | 0.76 |
ENST00000243457.4
|
KCNJ2
|
potassium inwardly rectifying channel subfamily J member 2 |
| chr1_+_25616780 | 0.76 |
ENST00000374332.9
|
MAN1C1
|
mannosidase alpha class 1C member 1 |
| chr16_+_4795378 | 0.75 |
ENST00000588606.5
|
SMIM22
|
small integral membrane protein 22 |
| chr6_-_31729260 | 0.73 |
ENST00000375789.7
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr16_+_4795357 | 0.70 |
ENST00000586005.6
|
SMIM22
|
small integral membrane protein 22 |
| chr8_-_80080816 | 0.69 |
ENST00000520527.5
ENST00000517427.5 ENST00000379097.7 ENST00000448733.3 |
TPD52
|
tumor protein D52 |
| chr6_-_31729478 | 0.69 |
ENST00000436437.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_-_33892010 | 0.68 |
ENST00000257818.3
|
LMO2
|
LIM domain only 2 |
| chr4_-_138242325 | 0.66 |
ENST00000280612.9
|
SLC7A11
|
solute carrier family 7 member 11 |
| chr2_-_177264686 | 0.62 |
ENST00000397062.8
ENST00000430047.1 |
NFE2L2
|
nuclear factor, erythroid 2 like 2 |
| chr17_+_63477052 | 0.62 |
ENST00000290866.10
ENST00000428043.5 |
ACE
|
angiotensin I converting enzyme |
| chr5_-_95284535 | 0.61 |
ENST00000515393.5
|
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr4_-_89836213 | 0.60 |
ENST00000618500.4
ENST00000508895.5 |
SNCA
|
synuclein alpha |
| chr6_-_31729785 | 0.58 |
ENST00000416410.6
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr6_-_10419638 | 0.58 |
ENST00000319516.8
|
TFAP2A
|
transcription factor AP-2 alpha |
| chr1_+_61077219 | 0.56 |
ENST00000407417.7
|
NFIA
|
nuclear factor I A |
| chr14_+_50533026 | 0.56 |
ENST00000441560.6
|
ATL1
|
atlastin GTPase 1 |
| chr22_-_37486357 | 0.55 |
ENST00000356998.8
ENST00000416983.7 ENST00000424765.2 |
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_+_135181323 | 0.54 |
ENST00000367814.8
|
MYB
|
MYB proto-oncogene, transcription factor |
| chr12_-_13095664 | 0.52 |
ENST00000337630.10
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr6_+_135181268 | 0.52 |
ENST00000341911.10
ENST00000442647.7 ENST00000618728.4 ENST00000316528.12 ENST00000616088.4 |
MYB
|
MYB proto-oncogene, transcription factor |
| chr2_+_169733811 | 0.51 |
ENST00000392647.7
|
KLHL23
|
kelch like family member 23 |
| chr1_-_16978276 | 0.50 |
ENST00000375534.7
|
MFAP2
|
microfibril associated protein 2 |
| chr7_-_120857124 | 0.50 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chr1_-_54887161 | 0.50 |
ENST00000535035.6
ENST00000371269.9 ENST00000436604.2 |
DHCR24
|
24-dehydrocholesterol reductase |
| chr7_-_151210488 | 0.50 |
ENST00000644661.2
|
H2BE1
|
H2B.E variant histone 1 |
| chr2_+_46297397 | 0.49 |
ENST00000263734.5
|
EPAS1
|
endothelial PAS domain protein 1 |
| chr6_+_17281341 | 0.49 |
ENST00000379052.10
|
RBM24
|
RNA binding motif protein 24 |
| chr6_-_33314202 | 0.48 |
ENST00000426633.6
ENST00000467025.1 |
TAPBP
|
TAP binding protein |
| chr17_-_58527980 | 0.45 |
ENST00000583114.5
|
SEPTIN4
|
septin 4 |
| chr13_+_31945826 | 0.43 |
ENST00000647500.1
|
FRY
|
FRY microtubule binding protein |
| chr9_-_22009272 | 0.42 |
ENST00000380142.5
ENST00000276925.7 |
CDKN2B
|
cyclin dependent kinase inhibitor 2B |
| chr2_-_24360299 | 0.42 |
ENST00000361999.7
|
ITSN2
|
intersectin 2 |
| chr7_+_20330893 | 0.42 |
ENST00000222573.5
|
ITGB8
|
integrin subunit beta 8 |
| chr6_-_37258110 | 0.41 |
ENST00000357219.4
ENST00000652386.1 ENST00000652639.1 |
TMEM217
|
transmembrane protein 217 |
| chr1_+_65792889 | 0.41 |
ENST00000341517.9
|
PDE4B
|
phosphodiesterase 4B |
| chr17_-_41168219 | 0.38 |
ENST00000391356.4
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr5_-_141878396 | 0.38 |
ENST00000503492.5
ENST00000287008.8 ENST00000394536.4 |
PCDH1
|
protocadherin 1 |
| chr5_+_76819022 | 0.37 |
ENST00000296677.5
|
F2RL1
|
F2R like trypsin receptor 1 |
| chr11_-_124762283 | 0.37 |
ENST00000444566.5
ENST00000278927.10 ENST00000435477.1 |
ESAM
|
endothelial cell adhesion molecule |
| chr1_-_15944348 | 0.37 |
ENST00000444358.1
|
ZBTB17
|
zinc finger and BTB domain containing 17 |
| chr2_+_64454145 | 0.37 |
ENST00000238875.10
|
LGALSL
|
galectin like |
| chr3_-_50337438 | 0.36 |
ENST00000327761.7
|
RASSF1
|
Ras association domain family member 1 |
| chr1_-_150697128 | 0.36 |
ENST00000427665.1
ENST00000271732.8 |
GOLPH3L
|
golgi phosphoprotein 3 like |
| chr16_-_46831134 | 0.36 |
ENST00000394806.6
ENST00000285697.9 |
C16orf87
|
chromosome 16 open reading frame 87 |
| chr19_+_55485176 | 0.35 |
ENST00000205194.5
ENST00000591590.1 ENST00000587400.1 |
NAT14
|
N-acetyltransferase 14 (putative) |
| chr18_+_58045683 | 0.35 |
ENST00000592846.5
ENST00000675801.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr7_-_100101333 | 0.34 |
ENST00000303887.10
|
MCM7
|
minichromosome maintenance complex component 7 |
| chr19_-_18280806 | 0.33 |
ENST00000600972.1
|
JUND
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr7_+_12687625 | 0.33 |
ENST00000651779.1
ENST00000404894.1 |
ARL4A
|
ADP ribosylation factor like GTPase 4A |
| chr19_-_42217667 | 0.32 |
ENST00000336034.8
ENST00000596251.6 ENST00000598200.1 ENST00000598727.5 |
DEDD2
|
death effector domain containing 2 |
| chr14_-_50532590 | 0.32 |
ENST00000013125.9
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr19_-_49423441 | 0.32 |
ENST00000270631.2
|
PTH2
|
parathyroid hormone 2 |
| chr15_+_88639009 | 0.32 |
ENST00000306072.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr17_+_41249687 | 0.31 |
ENST00000334109.3
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chr6_+_31815532 | 0.31 |
ENST00000375651.7
ENST00000608703.1 |
HSPA1A
|
heat shock protein family A (Hsp70) member 1A |
| chr5_+_57175803 | 0.31 |
ENST00000511209.5
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr19_+_11547840 | 0.30 |
ENST00000588935.1
|
CNN1
|
calponin 1 |
| chr2_-_174764407 | 0.30 |
ENST00000409219.5
ENST00000409542.5 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr11_+_66011994 | 0.30 |
ENST00000312134.3
|
CST6
|
cystatin E/M |
| chr18_+_58045642 | 0.30 |
ENST00000676223.1
ENST00000675147.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr11_-_66347560 | 0.30 |
ENST00000311181.5
|
B4GAT1
|
beta-1,4-glucuronyltransferase 1 |
| chr2_-_24360445 | 0.29 |
ENST00000443927.5
ENST00000406921.7 ENST00000412011.5 ENST00000355123.9 |
ITSN2
|
intersectin 2 |
| chr5_+_69492767 | 0.29 |
ENST00000681041.1
ENST00000680098.1 ENST00000680784.1 ENST00000396442.7 ENST00000681895.1 |
OCLN
|
occludin |
| chr1_-_154956086 | 0.28 |
ENST00000368463.8
ENST00000368460.7 ENST00000368465.5 |
PBXIP1
|
PBX homeobox interacting protein 1 |
| chr19_+_35248879 | 0.28 |
ENST00000347609.8
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr22_+_22811737 | 0.28 |
ENST00000390315.3
|
IGLV3-10
|
immunoglobulin lambda variable 3-10 |
| chr13_+_26254098 | 0.28 |
ENST00000381527.8
|
CDK8
|
cyclin dependent kinase 8 |
| chr15_+_88638947 | 0.27 |
ENST00000559876.2
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr2_-_29074515 | 0.27 |
ENST00000331664.6
|
PCARE
|
photoreceptor cilium actin regulator |
| chr7_-_100100716 | 0.27 |
ENST00000354230.7
ENST00000425308.5 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr7_-_93226449 | 0.26 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr12_+_32502114 | 0.26 |
ENST00000682739.1
ENST00000427716.7 ENST00000583694.2 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr8_-_92017292 | 0.26 |
ENST00000521553.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr17_+_29568591 | 0.26 |
ENST00000581411.6
ENST00000301057.8 |
TP53I13
|
tumor protein p53 inducible protein 13 |
| chr3_+_124584625 | 0.26 |
ENST00000291478.9
ENST00000682363.1 ENST00000454902.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr7_+_130380339 | 0.26 |
ENST00000481342.5
ENST00000604896.5 ENST00000011292.8 |
CPA1
|
carboxypeptidase A1 |
| chr12_-_79690522 | 0.25 |
ENST00000552637.1
|
PAWR
|
pro-apoptotic WT1 regulator |
| chr6_-_39322688 | 0.25 |
ENST00000437525.3
|
KCNK16
|
potassium two pore domain channel subfamily K member 16 |
| chr5_-_16936231 | 0.25 |
ENST00000507288.1
ENST00000274203.13 ENST00000513610.6 |
MYO10
|
myosin X |
| chr6_-_136550407 | 0.24 |
ENST00000354570.8
|
MAP7
|
microtubule associated protein 7 |
| chr1_-_156705764 | 0.24 |
ENST00000621784.4
ENST00000368220.1 |
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr17_-_58352139 | 0.23 |
ENST00000225504.8
|
SUPT4H1
|
SPT4 homolog, DSIF elongation factor subunit |
| chr1_-_156705575 | 0.23 |
ENST00000368222.8
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr19_+_35248728 | 0.23 |
ENST00000602003.1
ENST00000360798.7 ENST00000354900.7 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr19_+_35248694 | 0.23 |
ENST00000361790.7
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr1_+_155078829 | 0.23 |
ENST00000368408.4
|
EFNA3
|
ephrin A3 |
| chr17_+_9645502 | 0.22 |
ENST00000285199.12
|
USP43
|
ubiquitin specific peptidase 43 |
| chr7_+_131110087 | 0.22 |
ENST00000421797.6
|
MKLN1
|
muskelin 1 |
| chr2_-_219571529 | 0.22 |
ENST00000404537.6
|
OBSL1
|
obscurin like cytoskeletal adaptor 1 |
| chr15_+_75347030 | 0.22 |
ENST00000566313.5
ENST00000355059.9 ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei like DNA glycosylase 1 |
| chr11_+_111918900 | 0.22 |
ENST00000278601.6
|
C11orf52
|
chromosome 11 open reading frame 52 |
| chr7_-_130441136 | 0.22 |
ENST00000675596.1
ENST00000676312.1 |
CEP41
|
centrosomal protein 41 |
| chr19_+_35248656 | 0.22 |
ENST00000621372.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr13_+_41061481 | 0.22 |
ENST00000379487.5
|
WBP4
|
WW domain binding protein 4 |
| chr9_+_87497852 | 0.22 |
ENST00000408954.8
|
DAPK1
|
death associated protein kinase 1 |
| chr19_+_35248375 | 0.22 |
ENST00000602122.5
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr16_+_53820 | 0.22 |
ENST00000293861.8
ENST00000383018.7 ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein U11/U12 subunit 25 |
| chr1_-_17439657 | 0.22 |
ENST00000375436.9
|
RCC2
|
regulator of chromosome condensation 2 |
| chr3_+_50155024 | 0.22 |
ENST00000414301.5
ENST00000450338.5 ENST00000413852.5 |
SEMA3F
|
semaphorin 3F |
| chrX_-_30308333 | 0.22 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0 group B member 1 |
| chr3_-_48556785 | 0.22 |
ENST00000232375.8
ENST00000383734.6 ENST00000416568.5 ENST00000412035.5 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr7_-_85187039 | 0.22 |
ENST00000284136.11
|
SEMA3D
|
semaphorin 3D |
| chr16_+_283157 | 0.21 |
ENST00000219406.11
ENST00000404312.5 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A member 2 |
| chr11_+_68312542 | 0.21 |
ENST00000294304.12
|
LRP5
|
LDL receptor related protein 5 |
| chr9_-_131531178 | 0.21 |
ENST00000372210.7
ENST00000372211.7 |
UCK1
|
uridine-cytidine kinase 1 |
| chr14_-_20454741 | 0.21 |
ENST00000206542.9
ENST00000553640.3 |
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr12_-_75209701 | 0.21 |
ENST00000350228.6
ENST00000298972.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr12_-_75209422 | 0.21 |
ENST00000393288.2
ENST00000540018.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr5_-_151087131 | 0.21 |
ENST00000315050.11
ENST00000523338.5 ENST00000522100.5 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr5_-_160312524 | 0.21 |
ENST00000520748.1
ENST00000257536.13 ENST00000393977.7 |
CCNJL
|
cyclin J like |
| chr19_-_45768627 | 0.20 |
ENST00000560160.1
|
SIX5
|
SIX homeobox 5 |
| chr2_-_162152239 | 0.20 |
ENST00000418842.7
|
GCG
|
glucagon |
| chr6_+_33204645 | 0.20 |
ENST00000374662.4
|
HSD17B8
|
hydroxysteroid 17-beta dehydrogenase 8 |
| chr15_-_34336749 | 0.20 |
ENST00000397707.6
ENST00000560611.5 |
SLC12A6
|
solute carrier family 12 member 6 |
| chr1_-_45686501 | 0.20 |
ENST00000355105.8
ENST00000290795.7 |
GPBP1L1
|
GC-rich promoter binding protein 1 like 1 |
| chr19_-_50952942 | 0.19 |
ENST00000594846.1
ENST00000336334.8 |
KLK5
|
kallikrein related peptidase 5 |
| chr3_+_105367212 | 0.19 |
ENST00000472644.6
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr12_-_75209814 | 0.19 |
ENST00000549446.6
|
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr20_+_45416084 | 0.19 |
ENST00000279035.14
ENST00000543458.7 ENST00000372689.9 ENST00000639499.1 ENST00000279036.12 ENST00000638594.1 ENST00000638489.1 ENST00000638353.1 ENST00000639382.1 ENST00000640210.1 ENST00000639235.1 ENST00000638478.1 ENST00000432270.2 ENST00000545755.3 ENST00000640324.1 |
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T |
| chr14_-_57866075 | 0.19 |
ENST00000556826.6
|
SLC35F4
|
solute carrier family 35 member F4 |
| chr12_+_47963557 | 0.19 |
ENST00000256686.10
ENST00000549288.5 ENST00000429772.7 ENST00000552561.5 ENST00000546749.5 ENST00000552546.5 ENST00000550552.5 |
TMEM106C
|
transmembrane protein 106C |
| chr17_-_73092657 | 0.19 |
ENST00000580557.5
ENST00000579732.5 ENST00000578620.1 ENST00000542342.6 ENST00000255559.7 ENST00000579018.5 |
SLC39A11
|
solute carrier family 39 member 11 |
| chr13_-_20192928 | 0.19 |
ENST00000382848.5
|
GJB2
|
gap junction protein beta 2 |
| chr4_-_176195563 | 0.18 |
ENST00000280191.7
|
SPATA4
|
spermatogenesis associated 4 |
| chr19_-_39391137 | 0.18 |
ENST00000595564.5
|
PAF1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr22_+_49960760 | 0.18 |
ENST00000360612.5
|
PIM3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr10_+_102854251 | 0.18 |
ENST00000339834.10
ENST00000369883.3 |
BORCS7
|
BLOC-1 related complex subunit 7 |
| chr3_+_105366877 | 0.18 |
ENST00000306107.9
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr20_+_45416551 | 0.18 |
ENST00000639292.1
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis class T |
| chr5_-_160312756 | 0.18 |
ENST00000644313.1
|
CCNJL
|
cyclin J like |
| chr4_-_176269213 | 0.18 |
ENST00000296525.7
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr12_-_52834307 | 0.17 |
ENST00000330553.6
|
KRT79
|
keratin 79 |
| chr11_-_35418966 | 0.17 |
ENST00000531628.2
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr6_+_44216914 | 0.17 |
ENST00000573382.3
ENST00000576476.1 |
MYMX
|
myomixer, myoblast fusion factor |
| chr12_-_13095628 | 0.17 |
ENST00000457134.6
ENST00000537302.5 |
GSG1
|
germ cell associated 1 |
| chr3_-_51967410 | 0.17 |
ENST00000461554.6
ENST00000483411.5 ENST00000461544.2 ENST00000355852.6 |
PCBP4
|
poly(rC) binding protein 4 |
| chr19_+_50432885 | 0.17 |
ENST00000357701.6
|
MYBPC2
|
myosin binding protein C2 |
| chr3_+_127915226 | 0.17 |
ENST00000405109.5
|
KBTBD12
|
kelch repeat and BTB domain containing 12 |
| chr6_+_157981826 | 0.17 |
ENST00000355585.9
ENST00000640338.1 ENST00000367113.5 |
SYNJ2
|
synaptojanin 2 |
| chr11_-_35419098 | 0.17 |
ENST00000606205.6
ENST00000645303.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr7_+_50308672 | 0.17 |
ENST00000439701.2
ENST00000438033.5 ENST00000492782.6 |
IKZF1
|
IKAROS family zinc finger 1 |
| chr11_-_119381629 | 0.17 |
ENST00000260187.7
ENST00000455332.6 |
USP2
|
ubiquitin specific peptidase 2 |
| chr15_+_67254825 | 0.16 |
ENST00000629425.2
|
IQCH
|
IQ motif containing H |
| chr14_+_22271921 | 0.16 |
ENST00000390464.2
|
TRAV38-1
|
T cell receptor alpha variable 38-1 |
| chr11_+_44726811 | 0.16 |
ENST00000533202.5
ENST00000520358.7 ENST00000533080.5 ENST00000520999.6 |
TSPAN18
|
tetraspanin 18 |
| chr5_+_141489066 | 0.16 |
ENST00000252087.3
|
PCDHGC5
|
protocadherin gamma subfamily C, 5 |
| chr3_+_68006224 | 0.16 |
ENST00000496687.1
|
TAFA1
|
TAFA chemokine like family member 1 |
| chr11_-_44950151 | 0.16 |
ENST00000533940.5
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr6_-_39322540 | 0.16 |
ENST00000425054.6
ENST00000373227.8 ENST00000373229.9 |
KCNK16
|
potassium two pore domain channel subfamily K member 16 |
| chr19_-_43504711 | 0.16 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology like domain family B member 3 |
| chr11_-_35419462 | 0.16 |
ENST00000643522.1
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr6_+_108848387 | 0.16 |
ENST00000368972.7
ENST00000392644.9 |
ARMC2
|
armadillo repeat containing 2 |
| chr19_-_18522051 | 0.16 |
ENST00000262809.9
|
ELL
|
elongation factor for RNA polymerase II |
| chr18_-_55587335 | 0.16 |
ENST00000638154.3
|
TCF4
|
transcription factor 4 |
| chr14_-_106511856 | 0.16 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr6_+_135181361 | 0.15 |
ENST00000527615.5
ENST00000420123.6 ENST00000525369.5 ENST00000528774.5 ENST00000533624.5 ENST00000534044.5 ENST00000534121.5 |
MYB
|
MYB proto-oncogene, transcription factor |
| chr8_-_126557691 | 0.15 |
ENST00000652209.1
|
LRATD2
|
LRAT domain containing 2 |
| chr3_+_127915469 | 0.15 |
ENST00000407609.7
|
KBTBD12
|
kelch repeat and BTB domain containing 12 |
| chr4_+_5711154 | 0.15 |
ENST00000264956.11
ENST00000509451.1 |
EVC
|
EvC ciliary complex subunit 1 |
| chr4_-_139302516 | 0.15 |
ENST00000394228.5
ENST00000539387.5 |
NDUFC1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr2_-_164841410 | 0.15 |
ENST00000342193.8
ENST00000375458.6 |
COBLL1
|
cordon-bleu WH2 repeat protein like 1 |
| chr2_-_174764436 | 0.15 |
ENST00000409323.1
ENST00000261007.9 ENST00000348749.9 ENST00000672640.1 |
CHRNA1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chrX_+_71223216 | 0.15 |
ENST00000361726.7
|
GJB1
|
gap junction protein beta 1 |
| chr17_+_42833390 | 0.15 |
ENST00000590720.6
ENST00000586114.5 ENST00000585805.5 ENST00000441946.6 ENST00000591152.5 ENST00000589469.5 ENST00000293362.7 ENST00000592169.2 |
PSME3
|
proteasome activator subunit 3 |
| chr11_-_35419213 | 0.14 |
ENST00000642171.1
ENST00000644050.1 ENST00000643134.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr22_+_22343185 | 0.14 |
ENST00000427632.2
|
IGLV9-49
|
immunoglobulin lambda variable 9-49 |
| chr12_-_13095798 | 0.14 |
ENST00000396302.7
|
GSG1
|
germ cell associated 1 |
| chr3_+_52420955 | 0.14 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chr17_-_47957824 | 0.14 |
ENST00000300557.3
|
PRR15L
|
proline rich 15 like |
| chr1_-_154870264 | 0.14 |
ENST00000618040.4
ENST00000271915.9 |
KCNN3
|
potassium calcium-activated channel subfamily N member 3 |
| chr16_+_6483379 | 0.14 |
ENST00000552089.5
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr11_+_67586104 | 0.14 |
ENST00000495996.1
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr17_-_7241787 | 0.14 |
ENST00000577035.5
|
GABARAP
|
GABA type A receptor-associated protein |
| chr6_+_31971831 | 0.14 |
ENST00000375331.7
ENST00000375333.3 |
STK19
|
serine/threonine kinase 19 |
| chrX_+_111096211 | 0.13 |
ENST00000372010.5
ENST00000519681.5 |
PAK3
|
p21 (RAC1) activated kinase 3 |
| chr1_+_209768597 | 0.13 |
ENST00000487271.5
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr3_-_49813880 | 0.13 |
ENST00000333486.4
|
UBA7
|
ubiquitin like modifier activating enzyme 7 |
| chr4_+_168497066 | 0.13 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_+_168497044 | 0.13 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr13_-_41061373 | 0.13 |
ENST00000405737.2
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr6_+_41921491 | 0.13 |
ENST00000230340.9
|
BYSL
|
bystin like |
| chr6_+_150683593 | 0.13 |
ENST00000644968.1
|
PLEKHG1
|
pleckstrin homology and RhoGEF domain containing G1 |
| chr11_-_65862026 | 0.13 |
ENST00000532134.5
|
CFL1
|
cofilin 1 |
| chr17_-_41382298 | 0.13 |
ENST00000394001.3
|
KRT34
|
keratin 34 |
| chr18_-_72865680 | 0.12 |
ENST00000397929.5
|
NETO1
|
neuropilin and tolloid like 1 |
| chr19_+_7914823 | 0.12 |
ENST00000565886.2
|
TGFBR3L
|
transforming growth factor beta receptor 3 like |
| chr16_-_69385570 | 0.12 |
ENST00000566750.5
|
TERF2
|
telomeric repeat binding factor 2 |
| chrX_+_129738942 | 0.12 |
ENST00000371106.4
|
XPNPEP2
|
X-prolyl aminopeptidase 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.4 | 1.8 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.2 | 2.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 1.2 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.2 | 0.4 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.2 | 0.8 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 0.8 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 0.6 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.2 | 0.6 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 | 0.6 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.6 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 2.8 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 1.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.5 | GO:0050823 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.1 | 0.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.9 | GO:0014916 | tetrahydrobiopterin biosynthetic process(GO:0006729) regulation of lung blood pressure(GO:0014916) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.6 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.4 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.5 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.1 | 0.2 | GO:1903823 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.1 | 0.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.2 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.3 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0035732 | nitric oxide storage(GO:0035732) |
| 0.0 | 0.7 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.5 | GO:0048625 | norepinephrine metabolic process(GO:0042415) surfactant homeostasis(GO:0043129) myoblast fate commitment(GO:0048625) |
| 0.0 | 0.3 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.2 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.0 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0051511 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.2 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.2 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.7 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.3 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.3 | GO:0046959 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:1903892 | negative regulation of ATF6-mediated unfolded protein response(GO:1903892) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.7 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 4.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0097698 | telomere maintenance via base-excision repair(GO:0097698) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.3 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.4 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.3 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.0 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.4 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0003360 | brainstem development(GO:0003360) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 1.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 3.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 3.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.6 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.0 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 0.6 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 1.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.7 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.6 | GO:0070573 | tripeptidyl-peptidase activity(GO:0008240) peptidyl-dipeptidase activity(GO:0008241) bradykinin receptor binding(GO:0031711) metallodipeptidase activity(GO:0070573) |
| 0.1 | 1.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.6 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.5 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 0.9 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.1 | 0.2 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0035730 | S-nitrosoglutathione binding(GO:0035730) dinitrosyl-iron complex binding(GO:0035731) |
| 0.0 | 2.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.0 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.2 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 1.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |