|
chr4_+_73740541
Show fit
|
8.01 |
ENST00000401931.1
ENST00000307407.8
|
CXCL8
|
C-X-C motif chemokine ligand 8
|
|
chr1_+_183186238
Show fit
|
7.85 |
ENST00000493293.5
ENST00000264144.5
|
LAMC2
|
laminin subunit gamma 2
|
|
chr15_+_88638947
Show fit
|
7.71 |
ENST00000559876.2
|
ISG20
|
interferon stimulated exonuclease gene 20
|
|
chr11_-_102798148
Show fit
|
7.11 |
ENST00000315274.7
|
MMP1
|
matrix metallopeptidase 1
|
|
chr1_-_94541636
Show fit
|
7.09 |
ENST00000370207.4
|
F3
|
coagulation factor III, tissue factor
|
|
chr1_-_94541746
Show fit
|
7.05 |
ENST00000334047.12
|
F3
|
coagulation factor III, tissue factor
|
|
chr2_+_151357583
Show fit
|
6.79 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6
|
|
chr15_+_88639009
Show fit
|
6.60 |
ENST00000306072.10
|
ISG20
|
interferon stimulated exonuclease gene 20
|
|
chr1_-_153549120
Show fit
|
6.33 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3
|
|
chr1_-_153549238
Show fit
|
5.59 |
ENST00000368713.8
|
S100A3
|
S100 calcium binding protein A3
|
|
chr5_+_35856883
Show fit
|
4.98 |
ENST00000506850.5
ENST00000303115.8
ENST00000511982.1
|
IL7R
|
interleukin 7 receptor
|
|
chr18_+_63887698
Show fit
|
4.95 |
ENST00000457692.5
ENST00000299502.9
ENST00000413956.5
|
SERPINB2
|
serpin family B member 2
|
|
chr10_-_5978022
Show fit
|
4.49 |
ENST00000525219.6
|
IL15RA
|
interleukin 15 receptor subunit alpha
|
|
chr10_-_5977589
Show fit
|
4.40 |
ENST00000620345.4
ENST00000397251.7
ENST00000397248.6
ENST00000622442.4
ENST00000620865.4
|
IL15RA
|
interleukin 15 receptor subunit alpha
|
|
chr10_-_5977535
Show fit
|
4.35 |
ENST00000379977.8
|
IL15RA
|
interleukin 15 receptor subunit alpha
|
|
chr6_+_137867241
Show fit
|
4.11 |
ENST00000612899.5
ENST00000420009.5
|
TNFAIP3
|
TNF alpha induced protein 3
|
|
chr6_+_137867414
Show fit
|
4.10 |
ENST00000237289.8
ENST00000433680.1
|
TNFAIP3
|
TNF alpha induced protein 3
|
|
chr1_-_7940825
Show fit
|
4.04 |
ENST00000377507.8
|
TNFRSF9
|
TNF receptor superfamily member 9
|
|
chr10_+_102395693
Show fit
|
3.81 |
ENST00000652277.1
ENST00000189444.11
ENST00000661543.1
|
NFKB2
|
nuclear factor kappa B subunit 2
|
|
chr17_+_21288029
Show fit
|
3.60 |
ENST00000526076.6
ENST00000361818.9
ENST00000316920.10
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr6_-_44265541
Show fit
|
3.57 |
ENST00000619360.6
|
NFKBIE
|
NFKB inhibitor epsilon
|
|
chr12_-_104958268
Show fit
|
3.36 |
ENST00000432951.1
ENST00000258538.8
ENST00000415674.1
ENST00000424946.1
ENST00000433540.5
|
SLC41A2
|
solute carrier family 41 member 2
|
|
chr4_-_121164314
Show fit
|
3.32 |
ENST00000057513.8
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr1_+_156114700
Show fit
|
3.22 |
ENST00000677389.1
ENST00000368300.9
ENST00000368299.7
|
LMNA
|
lamin A/C
|
|
chr17_+_76385256
Show fit
|
3.14 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1
|
|
chr11_+_35176611
Show fit
|
3.10 |
ENST00000279452.10
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr1_+_156114251
Show fit
|
3.02 |
ENST00000361308.9
|
LMNA
|
lamin A/C
|
|
chr1_+_155135344
Show fit
|
3.01 |
ENST00000484157.5
|
SLC50A1
|
solute carrier family 50 member 1
|
|
chr21_-_35049238
Show fit
|
2.99 |
ENST00000416754.1
ENST00000437180.5
ENST00000455571.5
ENST00000675419.1
|
RUNX1
|
RUNX family transcription factor 1
|
|
chr10_-_5977492
Show fit
|
2.89 |
ENST00000530685.5
ENST00000397255.7
ENST00000379971.5
ENST00000528354.5
ENST00000397250.6
ENST00000429135.2
|
IL15RA
|
interleukin 15 receptor subunit alpha
|
|
chr21_-_35049327
Show fit
|
2.89 |
ENST00000300305.7
|
RUNX1
|
RUNX family transcription factor 1
|
|
chr11_+_35176696
Show fit
|
2.83 |
ENST00000528455.5
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr12_-_55842950
Show fit
|
2.79 |
ENST00000548629.5
|
MMP19
|
matrix metallopeptidase 19
|
|
chr5_+_136058849
Show fit
|
2.76 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced
|
|
chr11_-_65900375
Show fit
|
2.74 |
ENST00000312562.7
|
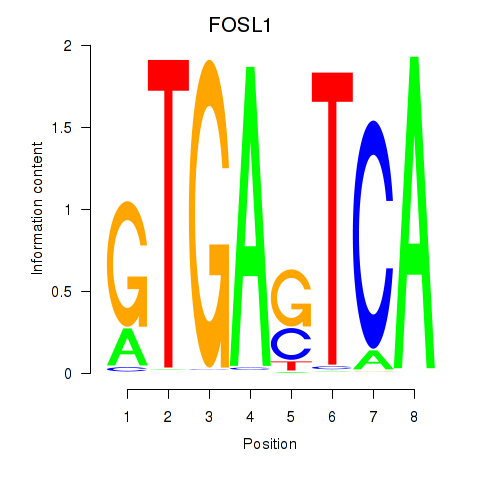
FOSL1
|
FOS like 1, AP-1 transcription factor subunit
|
|
chr11_-_65900413
Show fit
|
2.74 |
ENST00000448083.6
ENST00000531493.5
ENST00000532401.1
|
FOSL1
|
FOS like 1, AP-1 transcription factor subunit
|
|
chr11_-_65662780
Show fit
|
2.63 |
ENST00000534283.1
ENST00000527749.5
ENST00000533187.5
ENST00000525693.5
ENST00000534558.5
ENST00000532879.5
ENST00000406246.8
ENST00000532999.5
|
RELA
|
RELA proto-oncogene, NF-kB subunit
|
|
chr11_+_35176639
Show fit
|
2.60 |
ENST00000527889.6
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr11_+_35176575
Show fit
|
2.48 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr6_-_138107412
Show fit
|
2.43 |
ENST00000421351.4
|
PERP
|
p53 apoptosis effector related to PMP22
|
|
chr11_-_2929412
Show fit
|
2.42 |
ENST00000314222.5
|
PHLDA2
|
pleckstrin homology like domain family A member 2
|
|
chr8_+_125430333
Show fit
|
2.39 |
ENST00000311922.4
|
TRIB1
|
tribbles pseudokinase 1
|
|
chr1_-_209651291
Show fit
|
2.28 |
ENST00000391911.5
ENST00000415782.1
|
LAMB3
|
laminin subunit beta 3
|
|
chrX_-_155334580
Show fit
|
2.23 |
ENST00000369449.7
ENST00000321926.4
|
CLIC2
|
chloride intracellular channel 2
|
|
chr8_+_85245451
Show fit
|
2.15 |
ENST00000321764.4
|
CA13
|
carbonic anhydrase 13
|
|
chr18_+_58221535
Show fit
|
2.14 |
ENST00000431212.6
ENST00000586268.5
ENST00000587190.5
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr17_+_81683963
Show fit
|
2.10 |
ENST00000676462.1
ENST00000679336.1
ENST00000678196.1
ENST00000677243.1
ENST00000677044.1
ENST00000677109.1
ENST00000677484.1
ENST00000678105.1
ENST00000677209.1
ENST00000329138.9
ENST00000677225.1
ENST00000678866.1
ENST00000676729.1
ENST00000572392.2
ENST00000577012.1
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate
|
|
chr1_-_6602885
Show fit
|
1.99 |
ENST00000377663.3
|
KLHL21
|
kelch like family member 21
|
|
chr1_-_150235972
Show fit
|
1.98 |
ENST00000534220.1
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chr17_-_47851155
Show fit
|
1.91 |
ENST00000536300.2
|
SP6
|
Sp6 transcription factor
|
|
chr1_-_6602859
Show fit
|
1.86 |
ENST00000377658.8
|
KLHL21
|
kelch like family member 21
|
|
chr17_+_4950147
Show fit
|
1.86 |
ENST00000522301.5
|
ENO3
|
enolase 3
|
|
chr11_-_65663083
Show fit
|
1.85 |
ENST00000308639.13
|
RELA
|
RELA proto-oncogene, NF-kB subunit
|
|
chr7_+_143381286
Show fit
|
1.83 |
ENST00000449630.5
ENST00000322764.10
ENST00000457235.5
|
ZYX
|
zyxin
|
|
chr17_-_29930062
Show fit
|
1.81 |
ENST00000579954.1
ENST00000269033.7
ENST00000540801.6
ENST00000590153.1
ENST00000582084.1
|
SSH2
|
slingshot protein phosphatase 2
|
|
chr1_-_12616762
Show fit
|
1.76 |
ENST00000464917.5
|
DHRS3
|
dehydrogenase/reductase 3
|
|
chr16_+_57628507
Show fit
|
1.75 |
ENST00000456916.5
ENST00000567154.5
ENST00000388813.9
ENST00000562558.6
ENST00000566271.6
ENST00000563374.5
ENST00000673126.2
ENST00000562631.7
ENST00000568234.5
ENST00000565770.5
ENST00000566164.5
|
ADGRG1
|
adhesion G protein-coupled receptor G1
|
|
chr11_+_35618450
Show fit
|
1.72 |
ENST00000317811.6
|
FJX1
|
four-jointed box kinase 1
|
|
chr6_+_82363199
Show fit
|
1.72 |
ENST00000535040.4
|
TPBG
|
trophoblast glycoprotein
|
|
chr18_+_63970029
Show fit
|
1.71 |
ENST00000353706.6
ENST00000542677.5
ENST00000397985.7
ENST00000636430.1
ENST00000397988.7
ENST00000448851.5
|
SERPINB8
|
serpin family B member 8
|
|
chr7_+_28685968
Show fit
|
1.70 |
ENST00000396298.6
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr16_+_56608577
Show fit
|
1.66 |
ENST00000245185.6
ENST00000561491.1
|
MT2A
|
metallothionein 2A
|
|
chr10_+_24466487
Show fit
|
1.62 |
ENST00000396446.5
ENST00000396445.5
ENST00000376451.4
|
KIAA1217
|
KIAA1217
|
|
chr11_-_65662931
Show fit
|
1.61 |
ENST00000615805.4
ENST00000612991.4
|
RELA
|
RELA proto-oncogene, NF-kB subunit
|
|
chr10_-_25062279
Show fit
|
1.57 |
ENST00000615958.4
|
ENKUR
|
enkurin, TRPC channel interacting protein
|
|
chr2_+_102418642
Show fit
|
1.56 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr6_-_43629222
Show fit
|
1.51 |
ENST00000307126.10
|
GTPBP2
|
GTP binding protein 2
|
|
chr17_-_41140487
Show fit
|
1.49 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6
|
|
chr1_+_156126525
Show fit
|
1.49 |
ENST00000504687.6
ENST00000473598.6
|
LMNA
|
lamin A/C
|
|
chr9_+_35673917
Show fit
|
1.44 |
ENST00000617161.1
ENST00000378357.9
|
CA9
|
carbonic anhydrase 9
|
|
chr6_+_150721073
Show fit
|
1.43 |
ENST00000358517.6
|
PLEKHG1
|
pleckstrin homology and RhoGEF domain containing G1
|
|
chr18_+_58196736
Show fit
|
1.43 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr6_+_106098933
Show fit
|
1.41 |
ENST00000369089.3
|
PRDM1
|
PR/SET domain 1
|
|
chr12_-_214146
Show fit
|
1.37 |
ENST00000684302.1
|
SLC6A12
|
solute carrier family 6 member 12
|
|
chr6_-_46325641
Show fit
|
1.34 |
ENST00000330430.10
ENST00000405162.2
|
RCAN2
|
regulator of calcineurin 2
|
|
chr6_-_169250825
Show fit
|
1.28 |
ENST00000676869.1
ENST00000676760.1
|
THBS2
|
thrombospondin 2
|
|
chrX_-_111270474
Show fit
|
1.28 |
ENST00000324068.2
|
CAPN6
|
calpain 6
|
|
chr5_+_179820895
Show fit
|
1.28 |
ENST00000504627.1
ENST00000389805.9
ENST00000510187.5
|
SQSTM1
|
sequestosome 1
|
|
chr6_+_45328203
Show fit
|
1.26 |
ENST00000371432.7
ENST00000647337.2
ENST00000371438.5
|
RUNX2
|
RUNX family transcription factor 2
|
|
chr19_+_38304105
Show fit
|
1.26 |
ENST00000588605.5
ENST00000301246.10
|
C19orf33
|
chromosome 19 open reading frame 33
|
|
chr16_+_57628684
Show fit
|
1.23 |
ENST00000567397.5
ENST00000568979.5
ENST00000672974.1
|
ADGRG1
|
adhesion G protein-coupled receptor G1
|
|
chr10_-_30999469
Show fit
|
1.22 |
ENST00000538351.6
|
ZNF438
|
zinc finger protein 438
|
|
chr3_+_30606574
Show fit
|
1.21 |
ENST00000295754.10
ENST00000359013.4
|
TGFBR2
|
transforming growth factor beta receptor 2
|
|
chr21_-_26843012
Show fit
|
1.20 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1
|
|
chr18_-_49557
Show fit
|
1.19 |
ENST00000308911.8
|
TUBB8B
|
tubulin beta 8B
|
|
chr5_-_60488055
Show fit
|
1.16 |
ENST00000505507.6
ENST00000515835.2
ENST00000502484.6
|
PDE4D
|
phosphodiesterase 4D
|
|
chr17_+_76265332
Show fit
|
1.16 |
ENST00000327490.8
ENST00000587913.1
|
UBALD2
|
UBA like domain containing 2
|
|
chr1_+_150149819
Show fit
|
1.15 |
ENST00000369124.5
|
PLEKHO1
|
pleckstrin homology domain containing O1
|
|
chr21_-_26843063
Show fit
|
1.14 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1
|
|
chr12_+_6914571
Show fit
|
1.13 |
ENST00000229277.6
ENST00000538763.5
ENST00000545045.6
|
ENO2
|
enolase 2
|
|
chr1_-_150235943
Show fit
|
1.13 |
ENST00000533654.5
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chr6_-_83431038
Show fit
|
1.09 |
ENST00000369705.4
|
ME1
|
malic enzyme 1
|
|
chr19_+_35030711
Show fit
|
1.08 |
ENST00000595652.5
|
SCN1B
|
sodium voltage-gated channel beta subunit 1
|
|
chr12_-_55842927
Show fit
|
1.08 |
ENST00000322569.9
ENST00000409200.7
|
MMP19
|
matrix metallopeptidase 19
|
|
chr22_-_30246739
Show fit
|
1.07 |
ENST00000403987.3
ENST00000249075.4
|
LIF
|
LIF interleukin 6 family cytokine
|
|
chr1_-_112704921
Show fit
|
1.05 |
ENST00000414971.1
ENST00000534717.5
|
RHOC
|
ras homolog family member C
|
|
chr6_+_138161932
Show fit
|
1.05 |
ENST00000251691.5
|
ARFGEF3
|
ARFGEF family member 3
|
|
chr5_+_148203024
Show fit
|
1.04 |
ENST00000325630.3
|
SPINK6
|
serine peptidase inhibitor Kazal type 6
|
|
chr22_+_39520553
Show fit
|
1.03 |
ENST00000674920.3
ENST00000679776.1
ENST00000675582.2
ENST00000337304.2
ENST00000676346.2
ENST00000396680.3
ENST00000680446.1
ENST00000674568.2
ENST00000680748.1
ENST00000674835.2
|
ATF4
|
activating transcription factor 4
|
|
chr16_+_88453260
Show fit
|
1.03 |
ENST00000319555.8
|
ZFPM1
|
zinc finger protein, FOG family member 1
|
|
chr14_-_52791597
Show fit
|
1.01 |
ENST00000216410.8
ENST00000557604.1
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr1_+_156126160
Show fit
|
1.00 |
ENST00000448611.6
ENST00000368297.5
|
LMNA
|
lamin A/C
|
|
chr15_-_70096604
Show fit
|
0.99 |
ENST00000559048.5
ENST00000560939.5
ENST00000440567.7
ENST00000557907.5
ENST00000558379.5
ENST00000559929.5
|
TLE3
|
TLE family member 3, transcriptional corepressor
|
|
chr3_-_191282383
Show fit
|
0.98 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B
|
|
chrX_-_130110679
Show fit
|
0.97 |
ENST00000335997.11
|
ELF4
|
E74 like ETS transcription factor 4
|
|
chr4_-_165279679
Show fit
|
0.96 |
ENST00000505354.2
|
GK3P
|
glycerol kinase 3 pseudogene
|
|
chr22_+_20507571
Show fit
|
0.96 |
ENST00000414658.5
ENST00000432052.5
ENST00000292733.11
ENST00000263205.11
ENST00000406969.5
|
MED15
|
mediator complex subunit 15
|
|
chr3_-_37174578
Show fit
|
0.96 |
ENST00000336686.9
|
LRRFIP2
|
LRR binding FLII interacting protein 2
|
|
chr1_-_150235995
Show fit
|
0.95 |
ENST00000436748.6
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chr6_-_35921047
Show fit
|
0.93 |
ENST00000361690.7
ENST00000512445.5
|
SRPK1
|
SRSF protein kinase 1
|
|
chr11_-_76669985
Show fit
|
0.91 |
ENST00000407242.6
ENST00000421973.1
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr10_+_30434176
Show fit
|
0.91 |
ENST00000263056.6
ENST00000375322.2
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr8_-_69833338
Show fit
|
0.91 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1
|
|
chr1_-_28193873
Show fit
|
0.90 |
ENST00000305392.3
ENST00000539896.1
|
PTAFR
|
platelet activating factor receptor
|
|
chr7_+_128455849
Show fit
|
0.89 |
ENST00000435296.2
ENST00000257696.5
|
HILPDA
|
hypoxia inducible lipid droplet associated
|
|
chr8_-_27258414
Show fit
|
0.88 |
ENST00000523048.5
|
STMN4
|
stathmin 4
|
|
chr6_+_25652201
Show fit
|
0.88 |
ENST00000612225.4
ENST00000377961.3
|
SCGN
|
secretagogin, EF-hand calcium binding protein
|
|
chrX_-_49079702
Show fit
|
0.87 |
ENST00000636049.1
ENST00000474053.6
ENST00000635003.1
|
WDR45
|
WD repeat domain 45
|
|
chr10_+_30434021
Show fit
|
0.86 |
ENST00000542547.5
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr15_-_70096260
Show fit
|
0.85 |
ENST00000558201.5
|
TLE3
|
TLE family member 3, transcriptional corepressor
|
|
chr4_-_99657820
Show fit
|
0.85 |
ENST00000511828.2
|
C4orf54
|
chromosome 4 open reading frame 54
|
|
chr19_+_38390055
Show fit
|
0.85 |
ENST00000587947.5
ENST00000338502.8
|
SPRED3
|
sprouty related EVH1 domain containing 3
|
|
chr1_-_159924529
Show fit
|
0.84 |
ENST00000320307.8
|
TAGLN2
|
transgelin 2
|
|
chr16_-_31150058
Show fit
|
0.84 |
ENST00000569305.1
ENST00000268281.9
ENST00000418068.6
|
PRSS36
|
serine protease 36
|
|
chr10_-_49269
Show fit
|
0.83 |
ENST00000562809.1
ENST00000568866.5
ENST00000561967.1
ENST00000568584.6
|
TUBB8
|
tubulin beta 8 class VIII
|
|
chr7_+_148698857
Show fit
|
0.83 |
ENST00000663835.1
ENST00000655324.1
ENST00000662132.1
ENST00000666124.1
ENST00000325222.9
ENST00000660240.1
|
CUL1
|
cullin 1
|
|
chrX_-_130110479
Show fit
|
0.83 |
ENST00000308167.10
|
ELF4
|
E74 like ETS transcription factor 4
|
|
chr10_-_79949098
Show fit
|
0.82 |
ENST00000372292.8
|
SFTPD
|
surfactant protein D
|
|
chr18_+_58341038
Show fit
|
0.82 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr11_-_3840942
Show fit
|
0.81 |
ENST00000351018.5
|
RHOG
|
ras homolog family member G
|
|
chr19_-_45782388
Show fit
|
0.80 |
ENST00000458663.6
|
DMPK
|
DM1 protein kinase
|
|
chr11_-_9003994
Show fit
|
0.79 |
ENST00000309166.8
ENST00000525100.1
ENST00000531090.5
|
NRIP3
|
nuclear receptor interacting protein 3
|
|
chr6_-_35921079
Show fit
|
0.78 |
ENST00000507909.1
ENST00000373825.7
|
SRPK1
|
SRSF protein kinase 1
|
|
chr19_-_10339610
Show fit
|
0.78 |
ENST00000589261.5
ENST00000160262.10
ENST00000590569.1
ENST00000589580.1
ENST00000589249.1
|
ICAM3
|
intercellular adhesion molecule 3
|
|
chr19_-_45782479
Show fit
|
0.77 |
ENST00000447742.6
ENST00000354227.9
ENST00000291270.9
ENST00000683086.1
ENST00000343373.10
|
DMPK
|
DM1 protein kinase
|
|
chr2_+_68974573
Show fit
|
0.77 |
ENST00000673932.3
ENST00000377938.4
|
GKN1
|
gastrokine 1
|
|
chr19_+_35154715
Show fit
|
0.76 |
ENST00000392218.6
ENST00000543307.5
ENST00000392219.7
ENST00000541435.6
ENST00000590686.5
ENST00000342879.7
ENST00000588699.5
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr6_-_35921128
Show fit
|
0.75 |
ENST00000510290.5
ENST00000423325.6
|
SRPK1
|
SRSF protein kinase 1
|
|
chr1_-_27490130
Show fit
|
0.75 |
ENST00000618852.5
|
WASF2
|
WASP family member 2
|
|
chr9_-_35111573
Show fit
|
0.75 |
ENST00000378561.5
ENST00000603301.5
|
FAM214B
|
family with sequence similarity 214 member B
|
|
chr19_+_35154914
Show fit
|
0.74 |
ENST00000423817.7
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr3_-_11568764
Show fit
|
0.74 |
ENST00000424529.6
|
VGLL4
|
vestigial like family member 4
|
|
chr12_-_53232182
Show fit
|
0.72 |
ENST00000425354.7
ENST00000546717.1
ENST00000394426.5
|
RARG
|
retinoic acid receptor gamma
|
|
chr17_-_40755328
Show fit
|
0.71 |
ENST00000312150.5
|
KRT25
|
keratin 25
|
|
chr1_+_36155930
Show fit
|
0.70 |
ENST00000316156.8
|
MAP7D1
|
MAP7 domain containing 1
|
|
chrX_-_154019800
Show fit
|
0.69 |
ENST00000444230.5
ENST00000393687.6
ENST00000429936.6
ENST00000369974.6
ENST00000369980.8
|
IRAK1
|
interleukin 1 receptor associated kinase 1
|
|
chr9_-_35111423
Show fit
|
0.69 |
ENST00000378557.1
|
FAM214B
|
family with sequence similarity 214 member B
|
|
chr7_-_151024423
Show fit
|
0.69 |
ENST00000469530.4
ENST00000639579.1
|
ATG9B
|
autophagy related 9B
|
|
chr1_+_36156096
Show fit
|
0.68 |
ENST00000474796.2
ENST00000373150.8
ENST00000373151.6
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr9_-_107489754
Show fit
|
0.68 |
ENST00000610832.1
ENST00000374672.5
|
KLF4
|
Kruppel like factor 4
|
|
chr11_+_706117
Show fit
|
0.68 |
ENST00000533256.5
ENST00000614442.4
|
EPS8L2
|
EPS8 like 2
|
|
chr17_+_7351889
Show fit
|
0.66 |
ENST00000576980.2
|
KCTD11
|
potassium channel tetramerization domain containing 11
|
|
chr1_+_155033824
Show fit
|
0.64 |
ENST00000295542.6
ENST00000423025.6
ENST00000368419.2
|
DCST1
|
DC-STAMP domain containing 1
|
|
chrX_-_49079872
Show fit
|
0.64 |
ENST00000473974.5
ENST00000475880.6
ENST00000634838.1
ENST00000376372.9
ENST00000476728.5
ENST00000635666.1
ENST00000634559.1
ENST00000322995.13
ENST00000634736.1
ENST00000396681.9
ENST00000471338.6
ENST00000485908.6
ENST00000376368.7
ENST00000376358.4
|
WDR45
ENSG00000288053.1
|
WD repeat domain 45
novel protein
|
|
chr1_-_150236064
Show fit
|
0.63 |
ENST00000532744.2
ENST00000369114.9
ENST00000369115.3
ENST00000583931.6
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chr17_+_7352142
Show fit
|
0.63 |
ENST00000333751.8
|
KCTD11
|
potassium channel tetramerization domain containing 11
|
|
chr2_+_232697362
Show fit
|
0.63 |
ENST00000482666.5
ENST00000483164.5
ENST00000490229.5
ENST00000464805.5
ENST00000489328.1
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr9_-_120877026
Show fit
|
0.63 |
ENST00000436309.5
|
PHF19
|
PHD finger protein 19
|
|
chr1_-_207051202
Show fit
|
0.63 |
ENST00000315927.9
|
YOD1
|
YOD1 deubiquitinase
|
|
chrX_+_41333905
Show fit
|
0.62 |
ENST00000457138.7
ENST00000643821.1
|
DDX3X
|
DEAD-box helicase 3 X-linked
|
|
chr5_+_141489066
Show fit
|
0.62 |
ENST00000252087.3
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr11_+_35662739
Show fit
|
0.61 |
ENST00000299413.7
|
TRIM44
|
tripartite motif containing 44
|
|
chr3_+_48466222
Show fit
|
0.61 |
ENST00000625293.3
ENST00000492235.2
ENST00000635452.2
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1
|
|
chr15_-_48963912
Show fit
|
0.61 |
ENST00000332408.9
|
SHC4
|
SHC adaptor protein 4
|
|
chr2_+_169509693
Show fit
|
0.61 |
ENST00000284669.2
|
KLHL41
|
kelch like family member 41
|
|
chr2_-_179746040
Show fit
|
0.60 |
ENST00000409343.5
|
ZNF385B
|
zinc finger protein 385B
|
|
chr22_+_22030934
Show fit
|
0.59 |
ENST00000390282.2
|
IGLV4-69
|
immunoglobulin lambda variable 4-69
|
|
chr4_-_122621011
Show fit
|
0.57 |
ENST00000611104.2
ENST00000648588.1
|
IL21
|
interleukin 21
|
|
chr7_+_99408609
Show fit
|
0.56 |
ENST00000403633.6
|
BUD31
|
BUD31 homolog
|
|
chr6_+_30717433
Show fit
|
0.56 |
ENST00000681435.1
|
TUBB
|
tubulin beta class I
|
|
chr17_-_58517835
Show fit
|
0.56 |
ENST00000579921.1
ENST00000579925.5
ENST00000323456.9
|
MTMR4
|
myotubularin related protein 4
|
|
chr6_+_63521738
Show fit
|
0.56 |
ENST00000648894.1
ENST00000639568.2
|
PTP4A1
|
protein tyrosine phosphatase 4A1
|
|
chr9_+_4985227
Show fit
|
0.55 |
ENST00000381652.4
|
JAK2
|
Janus kinase 2
|
|
chr17_-_7687427
Show fit
|
0.55 |
ENST00000514944.5
ENST00000503591.1
ENST00000610292.4
ENST00000420246.6
ENST00000455263.6
ENST00000610538.4
ENST00000622645.4
ENST00000445888.6
ENST00000619485.4
ENST00000509690.5
ENST00000604348.5
ENST00000269305.9
ENST00000620739.4
|
TP53
|
tumor protein p53
|
|
chr10_+_122112957
Show fit
|
0.55 |
ENST00000369001.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2
|
|
chr11_+_706222
Show fit
|
0.55 |
ENST00000318562.13
ENST00000533500.5
|
EPS8L2
|
EPS8 like 2
|
|
chr4_-_36244438
Show fit
|
0.55 |
ENST00000303965.9
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr17_-_49210567
Show fit
|
0.54 |
ENST00000507680.6
|
GNGT2
|
G protein subunit gamma transducin 2
|
|
chr16_+_89921851
Show fit
|
0.53 |
ENST00000554444.5
ENST00000556565.5
|
TUBB3
|
tubulin beta 3 class III
|
|
chrX_-_49080066
Show fit
|
0.53 |
ENST00000634944.1
ENST00000423215.3
ENST00000465382.6
|
WDR45
|
WD repeat domain 45
|
|
chr21_-_42010327
Show fit
|
0.53 |
ENST00000398505.7
ENST00000449949.5
ENST00000310826.10
ENST00000398499.5
ENST00000398497.2
ENST00000398511.3
|
ZBTB21
|
zinc finger and BTB domain containing 21
|
|
chr1_-_150236150
Show fit
|
0.52 |
ENST00000629042.2
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E
|
|
chr15_-_22980334
Show fit
|
0.52 |
ENST00000610365.4
ENST00000617928.5
ENST00000611832.4
|
CYFIP1
|
cytoplasmic FMR1 interacting protein 1
|
|
chr16_+_83953232
Show fit
|
0.52 |
ENST00000565123.5
ENST00000393306.6
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr3_+_48465811
Show fit
|
0.52 |
ENST00000433541.1
ENST00000444177.1
|
TREX1
|
three prime repair exonuclease 1
|
|
chr17_+_49210699
Show fit
|
0.52 |
ENST00000225941.6
|
ABI3
|
ABI family member 3
|
|
chrX_+_49171889
Show fit
|
0.51 |
ENST00000376327.6
|
PLP2
|
proteolipid protein 2
|
|
chrX_-_49043345
Show fit
|
0.51 |
ENST00000315869.8
|
TFE3
|
transcription factor binding to IGHM enhancer 3
|
|
chr5_+_148202771
Show fit
|
0.51 |
ENST00000514389.5
ENST00000621437.4
|
SPINK6
|
serine peptidase inhibitor Kazal type 6
|
|
chr5_+_150497772
Show fit
|
0.51 |
ENST00000523767.5
|
NDST1
|
N-deacetylase and N-sulfotransferase 1
|
|
chr5_-_135578983
Show fit
|
0.50 |
ENST00000512158.6
|
CXCL14
|
C-X-C motif chemokine ligand 14
|
|
chr7_+_150368189
Show fit
|
0.49 |
ENST00000519397.1
ENST00000479668.5
|
REPIN1
|
replication initiator 1
|
|
chr17_-_29589536
Show fit
|
0.48 |
ENST00000394869.7
|
GIT1
|
GIT ArfGAP 1
|
|
chr5_-_138575359
Show fit
|
0.48 |
ENST00000297185.9
ENST00000678300.1
ENST00000677425.1
ENST00000677064.1
ENST00000507115.6
|
HSPA9
|
heat shock protein family A (Hsp70) member 9
|
|
chr1_-_109426410
Show fit
|
0.48 |
ENST00000271308.9
|
PSMA5
|
proteasome 20S subunit alpha 5
|
|
chr15_-_55917080
Show fit
|
0.47 |
ENST00000506154.1
|
NEDD4
|
NEDD4 E3 ubiquitin protein ligase
|
|
chr2_+_36696686
Show fit
|
0.47 |
ENST00000379242.7
ENST00000389975.7
|
VIT
|
vitrin
|
|
chr4_-_39032922
Show fit
|
0.47 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156
|
|
chr12_-_70788914
Show fit
|
0.47 |
ENST00000342084.8
|
PTPRR
|
protein tyrosine phosphatase receptor type R
|
|
chr19_+_49928702
Show fit
|
0.47 |
ENST00000595125.5
|
ATF5
|
activating transcription factor 5
|